Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences.2(1): 37–41Antimicrobial Activity of Ircinia sp; a Marine Sponge and Its Associated Bacteria from Andaman Coast
Pandian Krishnan, Suriamoorthy Veeramani, Sibnarayan Damroy, Kamal Sarma, Kumar Pavel, Jai Sunder*
*Corresponding author: [email protected]
ARTICLE CITATION: Krishnan P, Veeramani S, Damroy S, Sarma K, Pavel K and Sunder J (2014). Antimicrobial activity of Ircinia sp; a marine sponge and its associated bacteria from Andaman coast. Adv. Anim. Vet. Sci. 2 (1): 37 – 41.
Received: 2013–09–28, Revised: 2013–12–14, Accepted: 2013–12–16
The electronic version of this article is the complete one and can be found online at (http://dx.doi.org/10.14737/journal.aavs/2014.2.1.37.41) which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Abstract
Marine sponges and their surface associated bacteria are recognized as sink of many important bioactive compounds. A total of 28 bacterial isolates associated with the marine sponge, Ircinia sp. collected from North Bay, Andaman were screened for their antimicrobial activity against selected pathogens viz., Aeromonas hydrophila, Bacillus subtilis, Enterococcus durans, Streptococcus lentus, Klebsiella pneumoniae and Rolstonia solanacearum using disc diffusion assay. The bacterial isolates were identified based on their biochemical properties and were assigned to Vibrio sp., Aeromonas sp., Bacillus sp., Corynebacterium sp., Pseudomonas sp., Streptococcus sp., Enterococcus sp., Neisseria sp., Veillonella sp., Citrobacter sp. and Klebsiella sp. Of the total bacterial isolates tested, 66.67% were found to have antimicrobial properties ranging from broad spectrum to species specific. One of the isolates, I12 which was identified as Lysinibacillus fusiformis based on DNA sequence homology of 16S rDNA gene, inhibited Aeromonas hydrophila with about 120% of the efficiency of erythromycin. The present findings highlight the importance of sponge surface–associated bacteria in producing new natural compounds.
Introduction
The marine environment is an exceptional reservoir of bioactive natural products, many of which exhibit structural/chemical features not found in terrestrial natural products. A small number of marine plants, animals and microbes have already yielded more than 12,000 novel chemicals with hundreds of new compounds still being discovered every year (Donia and Hamann, 2003). To date majority of these chemicals have been identified from marine invertebrates of which sponges predominate (Lie and Zhou, 2002). More novel bioactive metabolites are obtained from sponges each year than from any other marine taxon and a range of pharmacological properties have been demonstrated (Blunt et al., 2006). Various ecological roles have also been proposed for these compounds, including defense against predators, competitors, fouling organisms and microbes. The toxic metabolites attributed to the sponges are most often produced by their associated bacteria (Piel et al., 2004). Symbiotic/epibiotic association between bacteria and marine sponges has been well known for a long time. To a sponge, different microbes can represent food sources, pathogens/parasites, or mutualistic symbionts. Several investigators (Boyd et al., 1999; Thakur et al., 2003, Manmadhan et al., 2005) have documented the possible role of associated bacteria in the epibacterial defense of the host. Sponges are known to maintain numerous bacteria which can amount to up to 40% of the biomass of a sponge. It has been reported that the ratio of bacteria with antimicrobial activity isolated from marine invertebrates was higher than that from other sources (Burgess et al., 1999). Antimicrobial activity has been reported for the bacteria associated with marine sponges collected from diverse areas viz., Antarctic region (Mangano et al., 2005), temperate (Chellossi et al., 2004) and tropical coastal waters (Thakur and Anil, 2000, Anand et al., 2006). Andaman and Nicobar group of Islands in India constitute about 25% of the country’s coast line and are bestowed with rich marine biodiversity. Sponges are one of the biologically and ecologically significant resources of the Islands which are hitherto unexplored for their pharmaceutical potential. Of the 486 sponge species reported from India (Thomas 1998) 61 are from Andaman. The number of sponge species reported from Andaman in the published reports does not by any scale represent the actual species richness as very few studies have been undertaken on investigating this important bioresource. Most of the sponge specimens reported from this region are those collected from the deep sea and there is no worthwhile study till date to document the sponge biodiversity in the shallow coastal waters from this group of islands. There are a few reports on antimicrobial properties of sponges from the Indian coast (Thakur and Anil, 2000; Anand et al., 2006; Gandhimathi et al., 2008), however, no report is available from the sponges collected from Andamans. The present study aims to characterize the cultivable heterotrophic fraction of surface–associated bacteria in a marine sponge, Ircinia sp. from Andaman; investigate their antimicrobial properties against selected pathogens and identify the isolates with significant bioactive potential using 16S rDNA sequence homology.
MATERIALS AND METHODS
Collection of Sponges
The marine sponge, Ircinia sp. (Figure 1) was collected using Self–Contained Underwater Breathing Apparatus (SCUBA) from a depth of 5–10 m in December 2008 at North Bay, located 5 km away from Port Blair. The sponge samples meant for the preparation of sponge extracts were rinsed with sterile seawater to remove adhering debris and associated biota and immediately placed in 1 L of Methanol (Merck, India). For isolating sponge associated bacteria, three sponge samples were placed separately in sample boxes containing pre–sterilized seawater, cooled in an insulation box, transported immediately to the laboratory and processed within 3 h. Ambient seawater was also collected in sterile glass bottles (Borosil, India) and brought to the laboratory in cold chain.
Isolation and Identification of Sponge Associated Bacteria (SAB)
After collection, the sponges were placed separately in beakers with sterile seawater, which were changed 4 to 5 times. One gram of sponge tissue was collected using a sterile knife and placed in a tube containing 10 ml of physiological saline (0.85% NaCl). After vigorous mixing, the samples were serially diluted up to 10–3 and 100 µl of each dilution was plated on Zobell marine agar plates. After incubating at 30°C for 48 h, the isolated colonies showing distinct morphology on agar plates were sub–cultured on a fresh agar plate. The isolated colonies were then streaked in slants in screw cap tubes and stored as pure cultures. A total of 62 isolates were obtained from the sponge surface, designated serially as S1–62 and stored for further screening. These bacterial isolates were identified to the genus level using Bergey’s Manual of Determinative Bacteriology (Buchanan and Gibbons, 1984). The biochemical test results were scored as 1 if positive or 0 if negative. The hierarchical cluster analysis was done using NTSYS 2.02 software. Clustering was performed by the unweighted pair–group method of analysis (UPGMA) with statistical support.
Preparation of Sponge Extract
The sponge sample was allowed to soak in methanol for 2 days with intermittent shaking after which the extract was decanted and collected. The extraction process was repeated 3 times, each time by adding equal volume of fresh methanol. These extracts were pooled, filtered and concentrated under reduced pressure using a rotary evaporator (BUCH1 Rotavapor R205, Switzerland). The crude sponge extract was then scrapped out and aliquots were made in phosphate buffered saline (PBS) at a concentration of 5 mg/ml. The sponge extract thus obtained was stored at 4°C till it was subjected to antibacterial activity tests.
Extraction of Bacterial Metabolites
A loop full of the pure cultures of the surface associated bacteria was inoculated in nutrient broth with 1.5% salt and incubated overnight in an orbital shaker (Orbitek, India). The broth was centrifuged (10,000 rpm for 10 min) when the bacterial growth reached the exponential phase at room temperature. The supernatant was collected, concentrated 10 fold in a vacuum drier, filter sterilized using a 45µ membrane filter (Millipore, USA) and stored at 4oC for subsequent screening of their antimicrobial properties.
Antibacterial Activity Assay
The sponge extracts and the bacterial metabolites were evaluated for their antibacterial activity against selected human, animal and plant pathogens viz., Aeromonas hydrophila, Enterococcus durans, Streptococcus lentus, Bacillus subtilis, Bacillus licheniformis, Streptococcus thermophilus, Klebsiella pneumoniae and Ralstonia solanacearum using disc diffusion assay (Hellio et al.,. 2001). A volume of 25 µl of the sponge extract and the concentrated supernatant from the sponge–associated bacterial isolates were used to saturate a sterile filter paper disc (Whatman, 6mm), allowed to dry at room temperature and placed on the surface of the marine agar plates freshly spread with 100 µl of the liquid cultures of each of the selected pathogens. The zone of inhibition (mm) was calculated from the edge of the disc after incubating the plates at 30oC for 24 h using standard antibiotic scale (Hi–Media, India). Discs soaked with same volume of methanol and sterile broths were used as negative controls, while those impregnated with antibiotics (Erythromycin, Ciprofloxacin; Hi–Media, India) were used as positive control. All the tests were performed in triplicate and the average zone of inhibition was calculated for each replicate assay of discs with sponge extracts / concentrated bacterial supernatants.
Phylogenetic Identification of Bacterial Isolates
The bacterial isolates were inoculated in LB broth and incubated at 30oC for 24 h. The bacterial culture was centrifuged at 10,000 rpm at RT for 10 min to collect the pellets, from which the genomic DNA was extracted by phenol–chloroform extraction method (Sambrook et al., 2001). The 16S rRNA gene was amplified from the bacterial genomic DNA using universal primers (Forward: PA 5’–AGA GTT TGA TCC TGG CTC AG –3’; Reverse: PH 5’–agg agg tga tcc agc cgca –3’). The PCR reaction mixture included 0.3 l of Taq polymerase, 2.5 l of Taq buffer, 2 l of dNTPs, 1 l each of forward and reverse primers, 1 l of genomic DNA and a final volume was made up to 25 l with DNAse free water (Fermentas, USA). Amplification by PCR was performed on a thermal cycler (Eppendorf, Germany) with a heated lid utilizing the conditions as follows: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 45 s, extension at 72°C for 90s; and a final extension at 72°C for 8 min. The size of the amplified product was verified by separating them on agarose gel electrophoresis (1%) against the 100bp ladder (Fermentas, USA). The 1500bp fragment of the 16S rRNA gene was then excised from the gel and purified using the QIAquick gel extraction kit (Qiagen, USA). The purified PCR products were sequenced from a commercial service provider (Bangalore Genei, India) using both forward and reverse primers.
Analysis of DNA Sequences
The 16S rRNA gene sequences obtained were compared with all other known rRNA sequences by performing a BLAST (Basic Local Alignment Search Tool) search (http:// www.ncbi.com/search BLAST.html). Multiple sequence alignment was performed with the similar sequences using the ClustalW program. The 16S rRNA gene sequences have been deposited into the GenBank database of NCBI (GenBank accession numbers: GQ487629 and GQ48763)
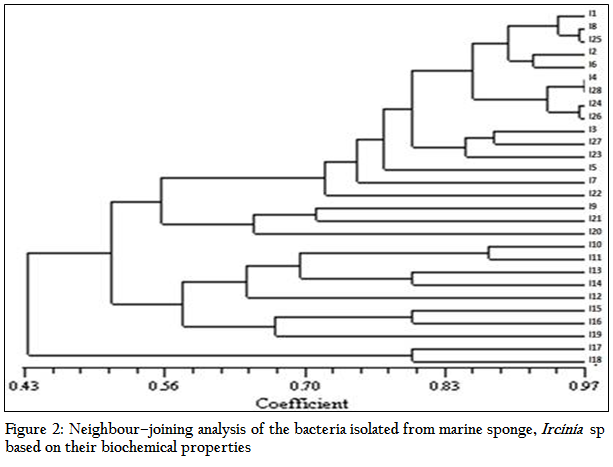
Figure 2: Neighbour–joining analysis of the bacteria isolated from marine sponge, Ircinia sp based on their biochemical properties
RESULTS AND DISCUSSION
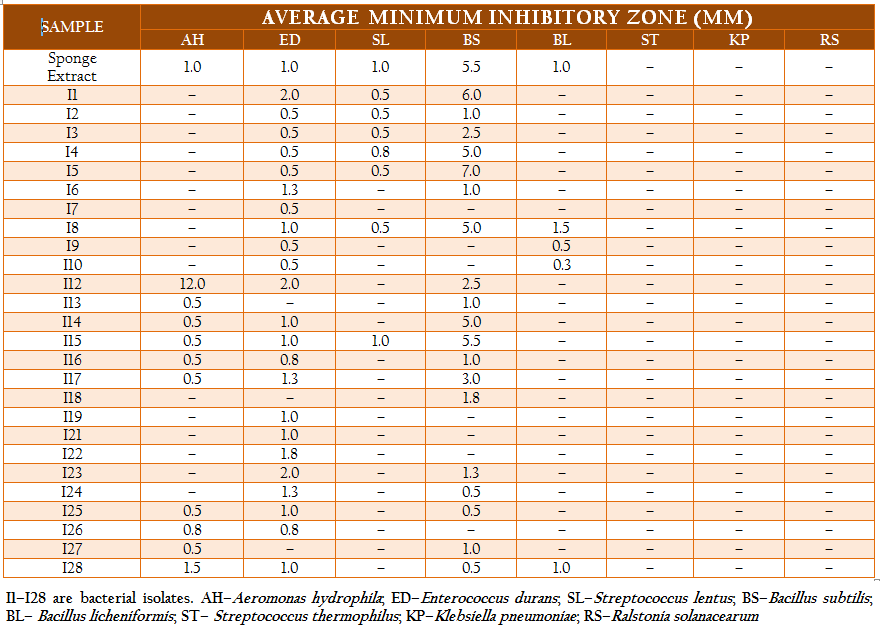
The total heterotrophic bacteria associated with the sponge, Ircinia sp. was found to be 5.8 x 106 cfu gm–2. A total of 71 bacterial strains which belonged to three different morphotypes were isolated. The majority were white circular (68%) followed by translucent circular (21%) and white irregular (11%) colonies. Based on their morphological, physiological and biochemical properties, the isolates were identified as Vibrio sp., Aeromonas sp., Bacillus sp., Corynebacterium sp., Pseudomonas sp., Streptococcus sp., Enterococcus sp., Neisseria sp., Veillonella sp., Citrobacter sp. and Klebsiella sp. The isolates were categorized and similarity percentages were studied based on their biochemical properties. Over 50% of the isolates were Gram positive and most of the isolates were motile (92%) and oxidase positive (85%). Carbohydrate utilization tests revealed that while most of the isolates could utilize glucose (90%), only 32% of them could utilize lactose or sucrose. The results of hierarchical cluster analysis based on the biochemical tests shows that at the similarity index of 75%, the isolates could be grouped into 11 clusters (Figure 2). A majority of the isolates (50%) belonged to group 1 while about 21% of the isolates were uniquely distinguished from the others.
The results of the antibacterial activity of the sponge extract and isolates obtained from Ircinia sp. are depicted in Table 1 and Figure 3. The sponge extracts and bacterial metabolites were tested for their antibacterial activity by disc diffusion method. Out of the total bacterial isolates, 26 were found to have antibacterial activity against various pathogens. The results revealed that the antibacterial activity was more pronounced against Gram positive bacteria (41.54%) than Gram negative bacteria (12.82%). There was significant (P<0.05) difference in the nature of inhibitory activity of the metabolites produced by different bacterial isolates. Majority of the bacterial metabolites (62.23%) showed narrow spectrum activities (against 2–3 organisms) against the tested organism. While 11.53% showed broad spectrum activities (against 4–5 organisms) and 19.23% showed activity against single pathogen. The antibacterial activity of the bacterial metabolite revealed that none of the metabolites and sponge extract exhibited activity against Streptococcus thermophilus, Klebsiella pneumoniae and Ralstonia solanacearam. Among the bacterial metabolites, highest antibacterial activity was showed against Enterobacter durans (88.46%) followed by Bacillus subtilis (73. 07%), Aeromonas hydrophilla (38.46%), Streptococcus lentus (26.92%) and Bacillus licheniformis (15.38%) respectively
The average zone of inhibition of the antibacterial activity of the sponge extract was recorded as 1.9 mm. Out of total bacterial metabolites which showed activity against various pathogens, seven bacterial isolates viz. I1, I4, I5, I8, I12, I14 and I15 showed better zone of inhibition than the sponge extract. The highest zone of inhibition was showed by I12 isolate against Aeromonas hydroiphilla (12 mm). Bacillus licheniformis was inhibited by only two isolates, I8–10 and I28.
Figure 3: Inhibitory efficiency of the extracts from Ircinia sp and its associated bacteria; AH–Aeromonas hydrophila; ED–Enterococcus durans; SL–Streptococcus lentus; BS–Bacillus subtilis; BL– Bacillus licheniformis
In the current study, samples of the marine sponge, Ircinia sp. collected from North Bay, South Andaman were collected. The bacterial isolates from Ircinia sp. belonged to the genera Vibrio sp., Aeromonas sp., Bacillus sp., Corynebacterium sp., Pseudomonas sp., Streptococcus sp., Enterococcus sp., Neisseria sp., Veillonella sp., Citrobacter sp. and Klebsiella sp. The study revealed great variation in the microbial ecology of the sponge surface bacteria as also reported earlier (Thakur et al., 2003) from different areas of the world. Gram positive bacteria generally predominate the marine environment (Bernen et al., 1997), while in some recent studies Gram positive and negative bacteria have been found in almost equal proportion (Anand et al.,2006). Gram positive bacteria constitute only 10% of the total isolates occurring in the Great Barrier Reef (Burja et al.,1999). However, in the present study, 60% of bacterial isolates associated with Ircinia sp. were found to be Gram positive. The flow chart exhibited distinct identification for me 8 and me 12. The I 8 isolates was depicted as Clostridium sp., where as I 12 was identified as Bacillus sp. (Anand et al., 2006) Earlier reports also suggested the prevalence of Pseudomonas, Bacillus and other species of bacteria on the surface of sponges (Thakur and Anil, 2000).
Various studies have been done on anti–microbial properties of the bacteria associated with the sponges. The antibiotics produce by this bacteria ranged from broad spectral to species specific (Anand et al., 2006). It is found that the antimicrobial activities of the bacteria associated with sponge protect the host sponges from various infections. Many sponge or sponge symbiont–derived metabolites are potent antibacterial, antifungal, antifeeding and antifouling compounds. In the present study also broad spectrum antibacterial activity was showed by both sponge extract and the bacterial metabolites. The methanol extract of Ircinia sp. had the highest inhibitory efficiency against B. subtilis. This could be attributed to the cumulative inhibitory efficiency of associated bacteria, as about 35% of the sponge isolates had significant (P < 0.05) antagonistic property against it.
The result of the present study showed the antibacterial activity against selected pathogens highlighting the role of associated bacteria. The bacterial load in the sponge was more than that in the surrounding water due to the ability of the sponges to concentrate bacteria in their body, as has been reported earlier (Friedrich et al., 2001). The inhibitory activity of isolates I1, I4, I5, I8, I12, I14 and I15 was significantly higher than that of the sponge extract, which attributes to the fact that the bacterial metabolites were obtained after enriching them in growth medium while the sponge extract simply had those isolates in densities as in nature. While all of the bacterial genera reported in the Ircinia sp. in the current investigation have been found to be associated with different organisms of the marine environment, their antimicrobial properties have not yet been documented. While seven of the isolates taken from Ircinia sp. have been found to have the cumulative inhibitory activity more than that of sponge extract, isolate I12 (Bacillus sp.) has been found to the most promising in terms of its antagonistic potential against the selected pathogens. The isolate I12 was identified as Bacillus sp. The 16S rDNA sequences of this isolate showed that they had 95% sequence homology with Lysinibacillus fusiforms. Kobayashi et al. (1994) characterized an antibiotic purified from a Vibrio sp. isolated from a marine sponge, Hyrtios altum. Radhika et al. (2007) isolated Cytophagus sp., Flexibacter sp., Pseudomonas sp. and Flavobacterium sp. from biofilms on the databuoys installed along the Indian coast.
The discovery of new classes of antibiotics is necessary due to the increased incidence of multiple resistances among pathogenic microorganisms to drugs that are currently in clinical use (Burgess et al., 1999). The marine sponges offer such great scope for exploration as they are the sink of many such bioactive compounds. The present investigation identified bacterial isolates associated with Ircinia sp., which contributed to the antagonistic property of the sponge from this geographically isolated and unique region. It underlines the inherent potential of exploring the marine sponges in Andamans for their bioactive properties.
ACKNOWLEDGEMENTS
Authors are grateful to the Director, CARI for providing financial assistance and facilities for carrying out the research work.
REFERENCES
Anand TP, Bhat AW, Shouche YS, Roy U, Siddhath J and Sarma SP (2006). Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of south east India. Microbiol. Res. 161:252–262. http://dx.doi.org/10.1016/j.micres.2005.09.002
Blunt JW, Copp BR, Munro MH, Northcote PT and Prinsep MR (2006). Marine natural products. Nat. Prod.Rep. 23:26–78. http://dx.doi.org/10.1039/b502792f
Boyd KG, Adams DR and Burgess JD (1999). Antibacterial and repellent activities of marine bacteria associated with algal surfaces. Biofoul. 14:227–236. http://dx.doi.org/10.1080/08927019909378414
Buchanan RE and Gibbons NE (1984). Bergey's manual of determinative bacteriology. 8th Edn. Science Publisher, Beijing.
Burgess JG, Jordan EM, Bregu M, Mearns–spragg A and Boyd KG (1999). Microbial antagonism: a neglected avenue of natural products research. J. Biotech. 70:27–32. http://dx.doi.org/10.1016/S0168-1656(99)00054-1
Burja AM, Webster NS, Murphy PT and Hill RT (1999). Microbial symbionts of Great Barrier Reef sponges. Mem. Queensland Mus. 44:63–67.
Donia M and Hamann MT (2003). Marine natural products and their potential applications as anti–infective agents. Lancet 3(6):338–348. http://dx.doi.org/10.1016/S1473-3099(03)00655-8
Hellio C, De la Broise D, Dufosse L, Lle Gal Y and Bourgougnon N (2001). Inhibition of marine bacteria by extracts of macroalgae: potential use for environmentally friendly antifouling paints. Mar. Environ. Res. 52:231–247. http://dx.doi.org/10.1016/S0141-1136(01)00092-7
Kobayashi M, Aoki S, Gato K, Matsunami K, Kurosu M and Kitagawa I (1994). Marine Natural Products. xxxiv. Trisindoline, a new antibiotic indole trimer, produced by a bacterium of Vibrio sp. separated from marine sponge Hyrtios altum. Chem.Pharm. Bull. 42:2449–2451. http://dx.doi.org/10.1248/cpb.42.2449
Lie J and Zhou J (2002). A marine natural product database. J. Chem. Inform. Model. 42(3): 742–748. http://dx.doi.org/10.1021/ci010111x
Mangano S, Michaaud L, Caruso C, Brilli M, Bruni V, Fani R and Giudice AL (2005). Antagonistic interactions between psychrotrophic cultivable bacteria isolated from antarctic sponges: a preliminary analysis. Res. in. Microbiol. 160:27–37. http://dx.doi.org/10.1016/j.resmic.2008.09.013
Manmadhan K, Sasaki H, Nakajima K, Nagata K and Nagata S (2005). Inhibitory activities of surface associated bacteria isolated from the marine sponge, Pseudoceratina purpurea. Microb. Environ. 20(3):178–185. http://dx.doi.org/10.1264/jsme2.20.178
Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N and Matsunaga S (2004). Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge, Theonella swinhoei. Proc. Nat. Acad. Sci. 101:16222–16227. http://dx.doi.org/10.1073/pnas.0405976101
Radhika G, Venkatesan R and Kathiroli S (2007). n–Methylpyrrolidone: isolation and characterization of the compound from the marine sponge, Clathria frondifera (Class: Demospongiae). Ind. J. Mar. Sci. 36:235–238.
Thakur NL and Anil AC (2000). Antibacterial activity of the sponge, Ircinia ramosa: importance of its surface associated bacteria. J. Chem. Ecol. 26:57–71. http://dx.doi.org/10.1023/A:1005485310488
Thakur NL, Hentschel U, Krasko A, Pabel CT, Anil AC and Müller W (2003). Antibacterial activity of the sponge, Suberites domuncula and its primorphs: potential basis for epibacterial chemical defense. Aqu. Microbio. Ecol. 31:77–83. http://dx.doi.org/10.3354/ame031077
Thomas PA (1998). Faunal diversity in India. A commemorative volume in the 50th years of India's independence. In: Alfred J.R.B. Das A.K. and Sanyal A.K. Editors. Zoological survey of India, Calcutta, p. 28–36.