Research Journal for Veterinary Practitioners
Research Article
Molecular and Phylogenetic Analysis of Canine Parvovirus Variants (2A-2B-2C) in Egypt
Wafaa M. El-Neshwy1, Heba El-Zahar2, Abdelkereem M. Morsi2, Tarek Shety2*
1Infectious Diseases, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Egypt; 2Internal Medicine, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Egypt.
Abstract | Canine parvovirus (CPV-2) infection is the main cause of fatal gastroenteritis in dogs. This study was directed to determine the variants of CPV-2 currently circulating in Egypt with reference to different risk factors that affecting occurrence of virus infection between dog populations. Seventy-eight dogs showed clinical signs of fever, different degrees of diarrhea, vomiting and anorexia out of total 117 dogs examined in different veterinary clinics at Sharkia and Dakahlia governorates during the period from February 2018 till June 2019. Using of CPV detection fast card test reported positive reaction in 48 fecal samples. PCR detection of canine parvovirus protein 2 (VP2) revealed positive reaction in 56 fecal swabs and 4 vomiting samples collected from clinically diseased dogs showing 360 bp bands by gel electrophoresis. Four positive fecal samples and one vomiting sample were randomly selected for sequencing. The fecal samples revealed presence of CPV-2a in one sample collected from Sharkia governorate and CPV-2b in three samples collected from Dakahlia governorate, while the vomiting sample collected from Dakahlia governorate showed CPV-2c. Protein alignment of the sequenced samples revealed mutation in different location of CPV-2b and CPV-2c. This is the first paper to detect CPV-2 (CPV-2c) antigen in the vomiting sample of naturally diseased dog by PCR. The highest rate of CPV-2 infection was found in the spring than other seasons, Pit bull dogs than other dog breeds, dogs aged from 2-4 months than other age groups and non-vaccinated than vaccinated dogs. We confirm that the three variants of CPV-2 (CPV-2a, CPV-2b and CPV-2c) are circulating in Egypt with rapid spread between the different governorates. CPV can be detected in the vomit of naturally infected dogs by PCR. Amino acid substitutions are continuously occurred in VP2 of CPV-2.
Keywords | Canine parvovirus, PCR, Sequencing, CPV 2C mutation, Risk factors.
Received | October 10, 2019; Accepted | December 23, 2019; Published | December 30, 2019
*Correspondence | Tarek Shety, Internal Medicine, Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Egypt; Email: [email protected]
Citation | El-Neshwy WM, El-Zahar H, Morsi AM, Shety T (2019). Molecular and phylogenetic analysis of canine parvovirus variants (2a-2b-2c) in Egypt. Res J. Vet. Pract. 7(3): 74-82.
DOI | http://dx.doi.org/10.17582/journal.rjvp/2019/7.3.74.82
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Shety et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Dogs are considered as the most popular pets reared by large scale of peoples in many countries. They present with different types and breeds that can be used for many purposes. Canine parvovirus (CPV) infection is one of the most common infectious diseases threatening the dog’s life. The disease causes severe hemorrhagic gastroenteritis (fever, vomiting, diarrhea and anorexia) (Macartney et al., 1984) in all ages especially dog with poor maternal immunity and poor vaccination regime (Folitse et al., 2018; Hueffer and Parrish, 2003; Yesilbag et al., 2007). In young puppies (4 - 6 weeks) the virus may cause myocarditis and death. Also, survivors of the acute viral myocarditis phase may show cardiomyopathy (Sime et al., 2015).
The virus replicates in the epithelial cell of the small intestine (intestinal villi) and sheds with huge numbers in the feces particularly 4 - 7 days post infection (Hoelzer et al., 2008), so feces of diseased dog serve as a main source of infection and fecal swab is the standard sample to be used for virus detection (Mylonakis et al., 2016).
CPV is a non-enveloped single strand DNA virus, genus
Table 1: Animals with signs of gastroenteritis from total examined dogs.
| Breed | Age groups | Sex | Vaccination state | Total | ||||||
| <2m | 2-4m | 4-6 m | 6m-1y | 1y-2y | Male | Female | Vaccinated | Unvaccinated and irregularly vaccinated | Clinically affected /total examined | |
| German shepherd | 3 /3 | 14 /21 | 2 /4 | 2/3 | 2 /5 | 17/22 | 6/14 | 7/16 | 16/20 | 23/36 |
| Pit bull | 0 | 9 /12 | 10/17 | 2/5 | 2 /3 | 11 /16 | 12 /21 | 7/20 | 16/17 | 23/37 |
| Husky | 0 | 9/11 | 2/4 | 0 | 1 /2 | 4/6 | 8 /11 | 2/6 | 10/11 | 12/17 |
| Mastiff | 0 | 1/2 | 0 | 0 | 5/5 | 1 /1 | 5/6 | 6/7 | 0/0 | 6/7 |
| Griffon | 0 | 7 /8 | 1 /4 | 0 | 0 | 3 /4 | 5 /8 | 3/5 | 5/7 | 8/12 |
| Rottweiler | 0 | 3 /5 | 0 | 0 | 0 | 1 /1 | 2 /4 | 2 /4 | 1/1 | 3/5 |
| Golden | 0 | 0 | 0 | 2 /2 | 1 /1 | 1 /1 | 2 /2 | 2/2 | 1/1 | 3/3 |
| Total | 3/3 |
43/59* |
15/29 | 6/10 | 11 /16 | 38/51 | 40/66 | 29/60 | 49/57 |
78/117 |
* significance at p<0.05
Protoparvovirus; family Parvoviridae (Appel et al., 1979). The virus is spherical shape (25nm in diameter) with total genome of about 5300 bp. The virus genome has two open reading frames; ORF1 which encodes two nonstructural proteins (NS1 and NS2) and ORF2 encodes two capsid proteins (VP1 and VP2) (Reed et al., 1988).
CPV is a variant of the feline panleukopenia virus (FPV) emerged in 1970 (Parrish et al., 1988b). The emerged virus acquired the ability to infect dog through some changes in its capsid protein. There are two types of the virus represented as CPV-1 (dog minute virus) and CPV-2 which are genetically and antigenically differ from each other (Cotmore et al., 2014). CPV-2 was recognized in 1978 (Burtonboy et al., 1979; Kelly, 1978). Due to mutation in five residues of VP2 during the period from 1979 to 1982, CPV-2 was replaced by CPV-2a that regained the ability to infect cat and other carnivores (Llamas-Saiz et al., 1996). The global replacement of CPV-2 by CPV-2a over a period of 2 to 3 years indicates that CPV-2a has a strong epidemiological advantage over CPV-2 (Parrish et al., 1988a). Further several mutations in VP2 of CPV-2a evolutes two new lineage of the virus (CPV-2b and CPV-2c). This can illustrate the important role of mutation occurred in VP2 in determining the antigenicity and host range of CPV. CPV-2b was first detected in the USA in 1984 while CPV-2c was identified in Italy in 2000 (Buonavoglia et al., 2001). Antigenic diversity between the three variants of CPV-2 depends mainly on point mutation in the genome especially at residue 426 of VP2. Presence of asparagine (Asn) amino acid (aa) at this residue evolutes CPV-2a but in CPV-2b and CPV-2c there is aspartic acid (Asp) and glutamic acid (Glu) in this residue respectively (Parrish et al., 1991).
In Egypt, the disease was first recorded in 1982 and recently was recorded by AL-Hosary (2018) and Soliman et al. (2018). They recorded that CPV-2a and CPV-2b were the circulating genotypes in Egypt but Awad et al. (2019) recorded only CPV-2b.
Canine parvovirus infection results in high morbidity may reach 100% and high mortality especially in puppies if no rapid diagnosis and treatment occurred (Appel et al., 1979). Rapid CPV Ag test kit is considered a helpful tool for rapid detection of virus infection. Also, PCR is recently a reliable method for detection of virus gene and differentiate the different variant of the virus (Schunck et al., 1995). The aim of the present study is to illustrate different lineages of CPV-2 circulating in Egypt with a first trial to detect the virus antigen in the vomit of the affected animals and clarifying the risk factors affecting the disease in Egypt.
Material and methods
Animals and Clinical Examination
One hundred and seventeen dogs from private veterinary clinics (Pet Care Clinic and Zoo Clinica) at Dakahlia governorate and (Alkawmia Pets Clinic and Veterinary Clinic of Faculty of Veterinary Medicine Zagazig University) at Sharkia governorate were included in the present study during the period from February 2018 to June 2019. The animals were from different breeds, ages, sex and vaccination state (depending on history taken from the owner) Table (1). Dogs were presented with signs of fever, diarrhea, vomiting and anorexia. All dogs were subjected to a unique standard clinical procedure included general signalment (age, sex and breed), history of the current illness, vaccination regime, general and specific clinical examination and clinical signs observation and recording according to
Table 2: Primers sequences, target genes, amplicon sizes and cycling conditions
| Target gene | Primers sequences | Amplified segment (bp) |
Primary denaturation |
Amplification (35 cycles) | Final extension | Reference | ||
| Secondary denaturation | Annealing | Extension | ||||||
| VP2 | Hfor: CAGGTGATGAATTTGCTACA | 630 |
94˚C 5 min. |
94˚C 30 sec. |
55˚C 40 sec. |
72˚C 45 sec. |
72˚C 10 min. |
Buonavoglia et al., 2001 |
| Hrev: CATTTGGATAAACTGGTGGT | ||||||||
Macartney et al. (1984). Fecal swabs and vomiting samples (if possible) were collected from dogs under investigation. Owners of the dogs have approved to include their dogs in the current study.
Sampling
Fecal swab were collected from all clinically diseased dogs and only four vomiting samples were collected from diseased dogs. The samples were used for detection of parvovirus antigen by the use of Rapid CPV Ag test kit (Zhenuri Biotech Co., Ltd., Shenzhen, China) and molecular detection of the virus by PCR. Collected samples were kept frozen at -70o C until analysis.
DNA Extraction
DNA extraction from samples was performed using the QIAamp DNA Mini kit (Qiagen, Germany, GmbH) with modifications from the manufacturer’s recommendations. Briefly, 200 µl of the sample suspension was incubated with 10 µl of proteinase K and 200 µl of lysis buffer at 56 oC for 10 min. After incubation, 200 µl of 100% ethanol was added to the lysate. The sample was then washed and centrifuged following the manufacturer’s recommendations. Nucleic acid was eluted with 100 µl of elution buffer provided in the kit. Oligonucleotide Primers, were supplied from Metabion (Germany) are listed in Table (2). PCR amplification, primers were utilized in a 25 µl reaction containing 12.5 µl of Emerald Amp Max PCR Master Mix (Takara, Japan), 1 µl of each primer of 20 pmol concentration, 7.5 µl of water, and 3 µl of DNA template. The reaction was performed in an Applied Biosystem 2720 thermal cycler.
Analysis of the PCR Products
The products of PCR were separated by electrophoresis on 1.5% agarose gel (Applichem, Germany, GmbH) in 1x TBE buffer at room temperature using gradients of 5V/cm. For gel analysis, 15 µl of the products was loaded in each gel slot. A gene ruler 100 bp ladder (Fermentas, Germany) and Gelpilot 100 bp ladder (Qiagen, Germany, GmbH) were used to determine the fragment sizes. The gel was photographed by a gel documentation system (Alpha Innotech, Biometra) and the data was analyzed through computer software.
DNA Sequencing and Phylogenetic Analysis
PCR products of randomly selected five tested positive samples (4 fecal swabs and one vomiting sample) were purified using QIAquick PCR product extraction kit (Qiagen, Valencia). Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer) was used for the sequence reaction and then it was purified using Centrisep spin column. DNA sequences were obtained by Applied Biosystem 3130 genetic analyzer (HITACHI, Japan); a BLAST® analysis (Basic Local Alignment Search Tool) (Altschul et al., 1990) was initially performed to establish sequence identity to Gen-Bank accessions. The sequence readings were assembled by DNA Baser 4.36.0.2 trial version and translated into protein sequences with the EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleotide and amino acid (AA) sequences were aligned using the CLUSTAL W method available in Clustal X 2.0 software.
The analyzed sequence and related protein of each sample was submitted on the Gen-Bank with accession number of (MN218609, MN218610, MN218611, MN218612, MN218613) for the five samples respectively. The phylogenetic tree was created by the MegAlign module of Laser gene DNA Star version 12.1 (Thompson et al., 1994) and phylogenetic analyses was done using maximum likelihood and neighbor joining in MEGA6 with using Canine Distemper partial sequence (AB025271) as an out group (Tamura et al., 2013). The translated amino acids of the five sequences were aligned with reference complete (CPV-2) and partial (CPV-2a, CPV-2b and CPV-2c) sequences on Gen-Bank using BioEdit 7.2.
Statistical Analysis
The data were analyzed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Chi-square test was used to study the effect of season, age and breeds on the results of different tests (rapid CPV test and PCR). P-value was set at p≤0.05.
Table 3: Prevalence of canine parvovirus infection in total examined dogs in different seasons.
| Season | |||||
| Spring | Summer | Autumn | Winter | Total | |
| 52 | 29 | 20 | 16 | 117 | |
| Clinical signs |
44 (84.62%)* |
4 (13.97%) | 14 (70%) | 16 (100%) | 78 (66.67%) |
| Rapid CPV Ag test kit |
32 (61.54%)* |
1 (3.45%) | 9 (45%) | 6 (37.5%) | 48 (41.03%) |
| PCR |
37 (71.15) * |
1 (3.45%) | 9 (45%) | 9 (56.25% | 56 (47.86%) |
* significance at p<0.05
Table 4: Dogs with positive PCR result for CPV-2 from clinically infected dogs.
| Breed | Age | Total | Vaccination state | Clinical signs | |||||||||
| <2m | 2-4m | 4-6 m | 6m-1y | 1y-2y | Total (%) | Vacci nated |
Un vaccinated |
Irregu larly vacc inated |
Bloody | Non bloody | Vom iting |
Death | |
|
German shepherd |
3 | 12 | 0 | 0 | 1 | 16 (28.57) | 3 | 10 | 3 | 7 | 9 | 15 | 4 |
| Pit bull | 0 | 8 | 8 | 2 | 0 | 18 (32.14) | 3 | 8 | 7 | 4 | 14 | 11 | 0 |
| Husky | 0 | 9 | 2 | 0 | 1 | 12 (21.43) | 2 | 5 | 5 | 6 | 6 | 10 | 5 |
| Mastiff | 0 | 0 | 0 | 0 | 1 | 1 (1.79) | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Griffon | 0 | 5 | 0 | 0 | 0 | 5 (8.93) | 0 | 3 | 2 | 4 | 1 | 5 | 0 |
| Rottweiler | 0 | 3 | 0 | 0 | 0 | 3 (5.36) | 2 | 1 | 0 | 1 | 2 | 3 | 0 |
| Golden | 0 | 0 | 0 | 0 | 1 | 1 (1.79) | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| Total | 3 | 37 | 10 | 2 | 4 | 56 (100) | 11 | 27 | 18 | 22 | 34 | 45 |
9 |
Table 5: Residue mutations present in protein sequences of studied samples, compared with gen-bank reference Complete (VP2 capsid protein) and partial sequence (CPV -2a, -2b and -2c).
| Accession No. | Residues | ||||||||||
| 267 | 270 | 297 | 300 | 307 | 324 | 367 | 370 | 426 | 440 | ||
| NP 955593 | Canine parvovirus | Phe | Cys | Ser | Ala | Asp | Tyr | Tyr | Gln | Asn | Thr |
| QDE54847 | CPV 2a | Tyr | * | Ala | Gly | Tyr | Ile | Asp | * | * | * |
| QDE54849 | CPV 2b | * | * | * | * | * | * | * | * | Asp | * |
| QDE54848 | Cpv 2C | * | * | * | * | * | * | * | Arg | Glu | * |
| MN218609 | CPV 2a | * | * | * | * | * | * | * | Gln | Asn | * |
| MN218610 | CPV 2b | * | * | * | * | * | * | * | * | Asp | Ala |
| MN218611 | CPV 2b | * | * | * | * | * | * | * | * | * | Ala |
| MN218612 | CPV2b | * | * | * | * | * | * | * | * | * | Ala |
| MN218613 | CPV2C | * | Ser | * | * | * | * | * | Arg | Glu | * |
Results
Clinical signs of gastroenteritis were observed in (78/117) examined dogs. Parvovirus infection was confirmed in 48 and 56 dogs by Rapid CPV Ag test kit and PCR respectively, from clinically infected dogs (78) Table (3). Diseased dogs showed lethargy, anorexia, dehydration, different degrees of diarrhea and vomiting. Nine dogs less than 3 months of age were died (Figure 1).
According to the results of statistical analysis, the highest rate of infection was reported in the spring (Temp. 35 - 40 oC and humidity 70 – 75 %) and the age group of 2 – 4 months of age. German shepherd, pit bull and husky dogs showed the highest rate of parvovirus infection than other species. Also, the risk of infection was higher in unvaccinated and irregularly vaccinated than vaccinated dogs (Table 3 and 4). No statistical difference was found in relation to sex while age has significant effect on the results where large proportion of the dogs were presented with signs of gastroenteritis due to CPV infection aged from 2-4 months (P<0.05).

Figure 1: a) Bloody watery diarrhea of CPV infected dog. b) Fluid therapy and blood transfusion for treatment of CPV infected Dogs. c) Husky dog suffering from lethargy and diarrhea due to CPV infection. d) Diagnosis of CPV infection in pit bull dog using Rapid CPV Ag test kit.
Molecular Detection of Canine Parvovirus
A band of about 360 pb was detected on agarose gel in 56 fecal samples and 4 vomiting samples from clinically infected dogs (Figure 2). It is the first trial to detect CPV form the vomit of the affected animal.
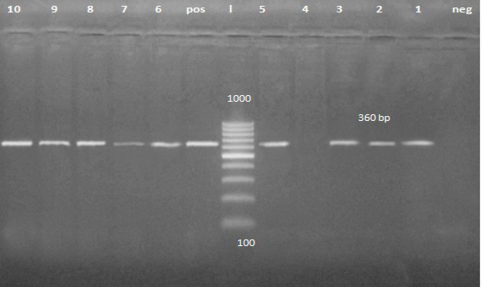
Figure 2: PCR product about 360 bp. Lane 1, 2, 3, 5, 6, 7, 8, 9 and 10 are positive samples while lane 4 is negative sample.
Primary analysis of the sequence of five randomly selected samples (4 fecal swabs and one vomiting sample) revealed presence of CPV-2a in one sample collected from Sharkia governorate and CPV-2b in three samples collected from Dakahlia governorate, while the vomiting sample collected from Dakahlia governorate showed CPV-2c.
Phylogenetic analysis based on VP2 of CPV-2 clustered the studied sequence of CPV-2a with CPV2a strains reported in India (KP071952) and in China (KF803597, KF803601 and KF803594). The three CPV-2b strains were clustered with CPV2b reported in Thailand (KP715689), in Egypt (KX358534) and in China (MH685921). While the studied CPV-2c strain was in one clade with CPV2c strains reported in china (MK460556 and MK460557) (Figure 3).
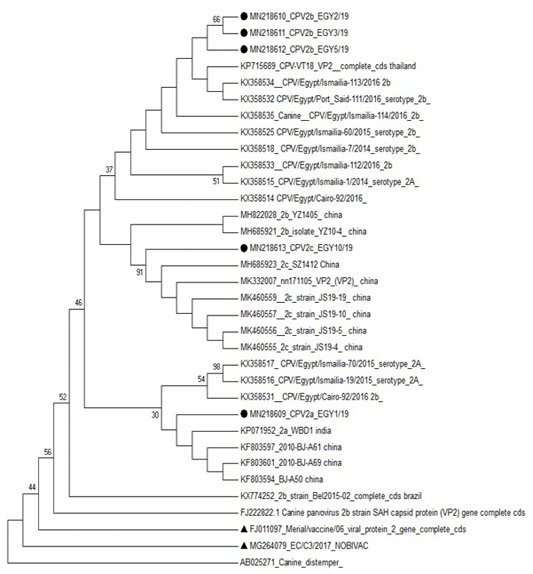
Figure 3: Phylogenetic analysis based on VP2 for the studied isolates against different sequences of canine parvovirus in Gen Bank. Studied samples were marked with dark circle and vaccine strains were marked with dark triangle.
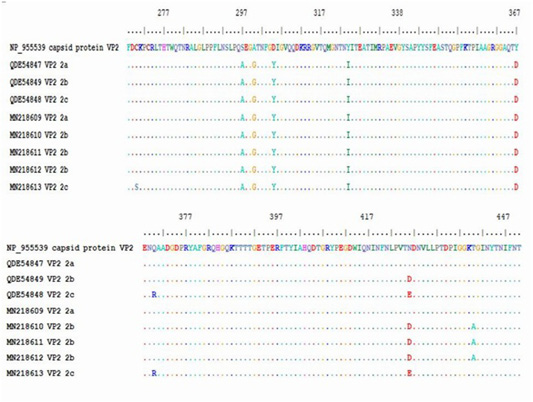
Figure 4: Multiple alignment of translated protein of studied variant (2a,2B,2C) against reference complete (VP2) and partial sequences in Gen-Bank.
The obtained sequence of each sample was translated into protein sequence with size of about 195aa starting at residue 258 until residue 452 of VP2 gene. The protein sequence was aligned with gen-bank reference complete (VP2 capsid protein) and partial sequence (CPV-2a, -2b and -2c). Residue 426 showed amino acid substitutions, (Asn426Asp) for CPV-2b and (Asn426Glu) for CPV-2c in the studied samples. All the five samples showed substitution of aa at residue 297 (Ser 297 Ala) which is characteristic of new CPV-2a, CPV-2b and CPV-2c beside the mutation occurred at 426 in each variant (Table 5).
The protein alignment showed mutation for the three CPV-2b studied samples at residue 440 (Thr 440 Ala). It is a first report of a mutation at residue 270 (Cys 270 Ser) for CPV-2c strain (Figure 4 and Table 5).
Discussion
Canine parvovirus causes a contagious gastroenteritis in dogs of all ages but mainly infect young puppies less than 6 months. This may be due to the high rate of mitosis in the intestinal crypt cells that provide a favorable environment for parvovirus replication (Cotmore et al., 2014). This interprets the higher rate of CPV-2 infection in puppies 2 - 4 months of age than other age groups recorded in this study.
Cases of infection reported in puppies less than 2 months of age may be due to poor maternal immunity supplied for this puppies or interference between maternal antibodies and vaccine strain. So, measuring the maternal antibodies is important before vaccination to avoid vaccination failure (Prittie, 2004).
Pit bull and German shepherd breeds appeared to be in risk of CPV-2 infection than other studied breeds. The unvaccinated or irregularly vaccinated dogs are more susceptible to infection than vaccinated ones. This came in agreement with previous results reported by (AL-Hosary, 2018; Kaur et al., 2014).
The highest risk of CPV-2 infection in dogs was recorded in Spring (71.15%) followed by Winter (56.25%) and Autumn (45%) this may attributed to the disturbance in climate which act as stress factor in animals. The etiology of hemorrhagic gastroenteritis is mainly attributed to the action of CPV-2 in the intestinal cell. It causes hyperemia, hemorrhage and atrophy of the intestinal mucosa. Necrosis, collapse of the lamina propria and villous atrophy are also reported by Oliveira et al. (2018).
PCR is high sensitive and specific technique for the identification of CPV-2 (Schunck et al., 1995). After 8-10 days of infection with CPV-2, specific antibodies in the intestinal lumen neutralize the virus and prevent its binding to cellular receptors and subsequent growth in cell cultures so PCR has special importance in this period (Buonavoglia et al., 2001).
In this study all CPV-2a, CPV-2b and CPV-2c were detected. Soliman et al. (2018) and Awad et al. (2019) recorded only CPV2b. Ali (2016) recorded CPV2c at Ismailia and Cairo. The recent variant CPV-2c was first recorded in this study at Dakahlia governorate. This means that the variant may be wide spread all over the country and further investigations are needed in the other governorates. Phylogenesis and blast analysis revealed 99.82% identity between the studied CPV-2c and the same variant recorded in china (MK268682). CPV-2a variant was 100% identical to the same variant detected in India (KP071952) (Dei Giudici et al., 2017). In Italy the new CPV-2b is the most wide spread variant while in Ecuador CPV-2a and CPV-2c showed the highest prevalence (De la Torre et al., 2018). CPV-2a is the predominant variant in Asia and Europe (Yi et al., 2016), CPV-2b is the predominant antigenic variant in African countries (Dogonyaro et al., 2013) while CPV-2c is thought to be the predominant variants in Latin America (Zhou et al., 2017).
In this study the puppy with CPV-2c showed severe gray watery diarrhea, vomiting, dehydration and anorexia. Similar signs were detected by Oliveira et al. (2018) in puppies infected with the same variant. They reported that bloody diarrhea is not common in CPV-2c infection although there was a high rate of mortality and infection of other organs of the body (lung, brain, liver and lymphoid tissues) in their study. These findings revealed that infection with CPV-2c is slightly differing from the common infection with CPV-2a and CPV-2b so accurate detection is necessary to avoid misdiagnosis with other causes of diarrhea.
This paper is the first to detect CPV-2c in the vomiting samples of naturally infected dog by PCR. Decaro et al. (2005) did not record any case of vomiting in CPV-2c infected dogs. They observed mild infection with rapid recovery. The contradictory result between the two studies may be explained by the environmental and breeding system difference, where the adult parent’s dogs in their study were regularly vaccinated while most animals used in our study were irregularly or not vaccinated. in our study PCR detected the nucleic acid of the virus in the vomit of diseased puppies but not infectious virus so presence of infectious virus in the vomit can only be hypothesized and vomit of infected dog may be a source of infection to susceptible one.
The peaks of the CPV capsid (three-fold spike domain) control cell tropism, host range and viral antigenicity through interaction with transferrin receptor (TFR) of the host cell (Strassheim et al., 1994). This region contain the residue 97 and 300 which are close to 426 and 297 residue of VP2 respectively (Hueffer and Parrish, 2003). Single mutation at the position 300 of VP2 is a key to determine whether a host is susceptible to virus infection or not. Glycine in this position makes the variant of CPV infective to dog cell. Thus, mutations in position 300 of VP2 play an important role in the cross-species transfer of viruses between different carnivores (Allison et al., 2016). Amino acid substitution of the three fold residues can also affect the ability of monoclonal neutralizing antibodies to bind the virus (Hafenstein et al., 2009; Llamas-Saiz et al., 1996).
The five sequenced samples had a substitution of aa at residue 297 (Ser 297 Ala). This substitution, along with the mutation in the 426 residue, lead to the designation “new CPV-2a” and “new CPV-2b (Decaro and Buonavoglia, 2012) and a new CPV-2c in this study.
The residue 440 of VP2 presents at the top of the threefold spike it may be the main antigenic site of the virus. The three CPV-2b variants in this study showed a mutation at this position (Thr 440 Ala) but CPV-2a and CPV-2c did not have mutation in this site. This type of mutation was detected for the first time by Battilani et al. (2002) in Italy another mutation at this position (Thr 440 Pro) was recorded by Dei Giudici et al. (2017).
Another mutation (Tyr 324 Ile) that present in the five used samples was previously detected by Perez et al. (2014) and Silva et al. (2017).
A recent mutation at 270 residues (Cys 270 ser) in CPV-2c was detected for the first time in the present study. Also the variant revealed another mutation at residue 370 (Gln 370 Arg).
The present study confirms that the CPV-2 has a high genetic mutation rate and the virus is still evolving. Although it is a DNA virus, it has high genetic substitution rate similar to RNA viruses (Decaro and Buonavoglia, 2012). Continuous accurate molecular epidemiological studies are needed to follow-up the new mutation of the virus genome which may result in vaccination failure. The continuous spread of new variants of the virus make it necessary to take care about the vaccines used against the diseases especially the available vaccines in Egypt are believed to be protective against all types of CPV-2.
Conclusion
CPV infection still threats dog’s life in spite of presence of available vaccine. The three variant of the virus are circulating in Egypt with continuous amino acid substitution in the virus genome so regular vaccination regime using an effective vaccine is very important for disease control. CPV-2 can be detected in the vomit of naturally infected dogs by PCR.
Acknowledgment
The authors express thanks to Dr. Shimaa Mohamed Galal, Professor of Virology, Faculty of Veterinary Medicine, Zagazig University, Egypt for her help in the virological aspects of this study. Great thanks for veterinarians and technicians in the reported veterinary clinics for their help in sampling and examination.
Conflict of interest
The authors declare that there is no conflict of interest.
Authors Contribution
All authors contributed equally to this work. They designed the study, collected the samples, analyzed the data and wrote the manuscript. All the authors have read and approved the final draft of the manuscript.
References






