The Journal of Advances in Parasitology
Short Communication
First Report on Molecular Detection of Toxocara Species of Dogs in Pakistan
Suleman Shah1, Haroon Akbar1, Nisar Ahmad1, Raheela Akhtar2*, Wasim Shehzad3, Saher Islam3
1Department of Parasitology, University of Veterinary and Animal Sciences, Lahore, Pakistan. 54000; 2Department of Pathology, University of Veterinary and Animal Sciences, Lahore, Pakistan. 54000; 3Institute of Biochemistry and Biotechnology, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Abstract | The present study was designed to provide the information about molecular detection of Toxocara species in dogs of Pakistan for the first time. One hundred fecal samples from suspected dogs were collected from Pet Centre, University of Veterinary and Animal Sciences, Lahore and screened for Toxocara eggs through microscopy. The microscopically positive samples were further subjected to Polymerase chain reaction (PCR) targeting internal transcribed spacer region of Toxocara genome. The PCR positive samples were further purified and sequenced in both directions using Sanger sequencing method. The results indicated three samples (1.5%) positive for Toxocara through fecal microscopy while sequencing confirmed these entire samples positive for T. leonina. This study precisely diagnosed Toxocara leonine for the first time in dogs to nullify the chance of misdiagnosis by conventional fecal microscopy.
Keywords | Dog, Polymerase chain reaction, Sequencing, Toxocara leonina
Editor | Muhammad Imran Rashid, Department of Parasitology, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | March 13, 2018; Accepted | April 03, 2018; Published | June 25, 2018
*Correspondence | Raheela Akhtar, Department of Pathology, University of Veterinary and Animal Sciences, Lahore, Pakistan; Email: [email protected]
Citation | Shah S, Akbar H, Ahmad N, Akhtar R, Shehzad W, Islam S (2018). First report on molecular detection of toxocara species of dogs in Pakistan. J. Adv. Parasitol. 5(2): 25-28.
DOI | http://dx.doi.org/10.17582/journal.jap/2018/5.2.25.28
Copyright © 2018 Shah et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Toxocara are worldly distributed zoonotic nematodes of dogs and cats mainly including Toxocara canis, Toxocara cati and Toxocara leonina (Le et al., 2016). Various diagnostic methods are employed for the identification of Toxocara such as fecal microscopy and immunodiagnostic techniques. However, these methods have their inherent limitations. Coproscopical techniques are classical and are extensively used to identify eggs and larvae in fecal samples but cannot differentiate different species of Toxocara such as Toxocara canis and T. cati by only measuring the size of eggs. As both species have almost same egg shape and size this may prone to inaccurate diagnosis (Jimenez et al., 2016).
Similarly, Enzyme linked immunosorbent assay (ELISA) have also been employed by using excretory/secretory antigens of larvae but cross reactivity of these antigens does not discriminate between different species of Toxocara and thus cannot ensure the accurate diagnosis particularly in mixed infection (Medawar et al., 2017).
Alternatively, the DNA based molecular approaches such PCR have been introduced for confirmatory diagnosis of toxocariosis at species level. This may overcome the drawbacks of traditional methodologies due to high sensitivity and specificity of PCR. The development and use of such methods can effectively control toxocara infection as PCR permits specific amplification of a small amount of genomic DNA, for example of a single Toxocara or its egg (Gasser et al., 2013).
Keeping in view the above mentioned facts the present study was designed to achieve the goal of accurate diagnosis of prevalent Toxocara species of dogs in Pakistan using PCR and DNA sequencing. As DNA based studies had never been employed for the diagnosis of toxocariasis in Pakistan before, this adds a novel aspect to current study.
MATERIALS AND METHODS
One hundred fecal samples of dogs were collected from Pet Centre, University of Veterinary & Animal Sciences, Lahore and were examined through flotation technique to observe the presence of ova of Toxocara species by using Sheather’s flotation solution. The samples positive for Toxocara were stored into 70% ethanol.
Molecular Detection of Toxocara Species
DNA was extracted from fecal samples using TIANGEN stool kit (Tiangen, Beijing, China). The extraction protocol was modified for cell lysis by overnight incubation of egg suspension at 60°C. 200µL egg suspension was mixed with 1.4mL of GSL and vortexed for one minute to homogenize the sample. The suspension was incubated overnight at 60°C and vortexed again for 15 seconds and centrifuged at 12000rpm for two minutes. Supernatant (1.2 mL) was transferred into a new micro centrifuge tube and one inhibitex tablet was added to each sample and vortexed thoroughly for the absorption of inhibitors. This mixture was again centrifuged at 12000rpm for three minutes and the supernatant was mixed with 15µL protinease K and 200µL lysis buffer and vortexed for 15 seconds. The sample was incubated at 70°C for 10 minutes to yield a homogenous mixture. Absolute ethanol (200 µL) was added to the sample and mixed thoroughly by vortex for 15 seconds. The solution was pipetted in a spin column placed in a collection tube centrifuged at 12000 rpm for 30 seconds and the flow was discarded. 500µL wash buffer was added to the spin column and centrifuged at 12000 rpm for 30 seconds again. The flow was discarded. 700μL buffer PW was added and centrifuged at 12000rpm for 30 seconds. The flow was discarded and the spin column was put in the collection tube. 500μL elution buffer was added to TIANamp spin column CR2, centrifuged at 12,000 rpm for 30 seconds, the flow was discarded and the spin column was centrifuged at 12000 rpm for two minutes to dry the membrane completely. The spin column was put in a new 1.5mL micro centrifuge tube and 50µL distilled water was added directly to the center of the membrane incubated at room temperature for five minutes and centrifuged at 12000 rpm for two minutes the spin column was discarded and the DNA sample was stored at 4°C for further analysis. The extracted DNA samples were dissolved in low T.E buffer and working aliquots were made while stock DNA was stored at -20°C.
Designing of Primers
We used MegAlign program of DNASTAR Lasergene software for the alignment of ITS-2 sequences from Genebank of T. canis, T. cati and T. leonine. The consensus sequence was generated by Clustal W analysis of aligned sequences. A set of primers was designed by PrimerSelect program of DNASTAR Lasergene (DNAstar, Madison, WI) and used to amplify ITS-2 region of Toxocara species.
PCR conditions were optimized for the amplification of Toxocara internal transcribed spacer-2 (ITS-2) region of
|
Primer Pair |
Toxocara General Primer Forward | TTGAGGGGTTTTGGGTGAGC |
| Toxocara General Primer Reverse | GCATTTTCTAGGTGAGGCGGA |
rRNA gene. Initial denaturation was at 94°C for two minutes followed by the denaturation at same temperature for 30 seconds. Annealing was done at 60°C for 45 seconds. After annealing two extension steps were carried out at 72°C for 45 seconds and 10 minutes consecutively. PCR products were visualized on 2% agarose gel using gel documentation system. The selected samples were purified using gel extraction kit (Invitrogen).
Sequencing of PCR Amplicons
Amplified DNA samples were sequenced by Sanger sequencing method at 1st BASE DNA Sequencing Services (Singapore). Sequences of Internal Transcribed Sequence (ITS) from Genbank of National Center for Biotechnology Information (NCBI) Bethesda were retrieved. They were aligned using CLUSTAL W to quantify genetic distances among isolates Toxocara spp. in dogs. Geneious R8.1.6 software was used for phylogenetic and Bootstrapping analysis. The P (uncorrected distance often referred as p-distances or dissimilarity distance) and Jukes–Cantor distances were used to construct a neighbor-joining tree. We applied parsimony method with bootstrapping (1000 replicates). We used subunit ribosomal RNA gene of Trichinella papuae (AY851286) for genotype analysis of Toxocara species (Van et al., 2013). The sequencing results were subjected to blast using Basic Local Alignment Search Tool (BLAST) provided by NCBI. The results obtained from fecal microscopy were subjected to statistical analysis applying Pearson Chi-Square test using SPSS software version 20.0.
RESULTS AND DISCUSSION
Diagnosis of toxocariosis largely depends upon conventional microscopy and serological techniques. However, these techniques are not very much reliable due to low specificity and sensitivity. The present study showed that the genetic markers in the ITS-2 region of Toxocara DNA could be used for the detection of Toxocara species by employing PCR.
Out of 100 fecal samples 26 were microscopically positive for Toxocara eggs (Figure 1) while only three (1.5%) were PCR positive. The microscopy results in present study coincide with previous studies of Chattha et al. (2009) who also reported 30% and 49% incidence in pet and stray dogs in Lahore by microscopy but our PCR results clearly indicate an increased number of false positive cases by microscopy due to closely related morphology of eggs in present
and past studies. These results surely highlight the need for molecular confirmation.
The age wise occurrence of Toxocara was 3.1% in (0 to 6 months), 5.1% in (6 to 12 months) and no animal positive for Toxocara infection in (above 12 months). As there was a non-significant difference in incidence at various age groups (P≥0.05) this showed that there was no relationship between Toxocara infection and age of dogs. This is not in agreement with the studies of Shabbir et al. (2010) who found more incidences of Toxocara canis and Toxocara cati in young puppies and kittens.
An amplicon of 520bp was amplified from three samples positive for fecal samples (Figure 2).
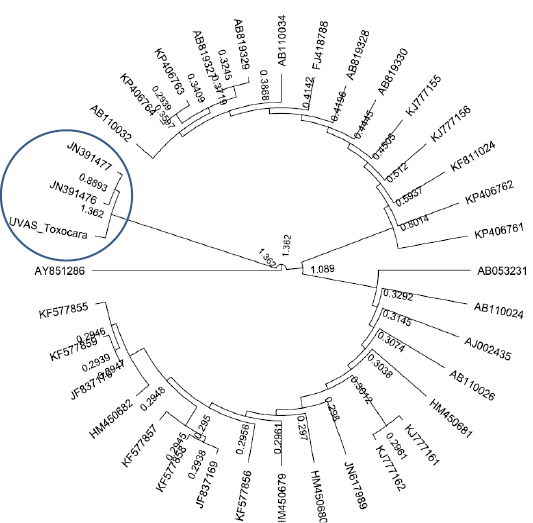
All purified samples were subjected to DNA sequencing in both directions. The sequencing results showed homology with Toxocara leonine in all three samples. This was a unique finding as Toxocara leonine has not been reported before in dogs of Pakistan. The Phylogenetic analyses showed the relevance our sequence to Toxascaris leonina (JN391477and JN391477) as shown in Figure 3.
Conclusions
Toxocara leonina was first time reported in dogs of Pakistan as this specie also has zoonotic importance therefore the results of present study can be implemented to prevent disease spread in humans and adopt control policies in animals. As all the previous studies of toxocariasis in Pakistan are serology based (Shabbir et al., 2010; Ahmad et al., 2011) and this was the first successful attempt of using PCR for detection of Toxocara species in Pakistan therefore this technique can be further applied in future for confirmatory and routine diagnosis of toxocariasis in dogs and humans.
Acknowledgements
This study was funded by Grand Challenges Canada project No. S4_0266-01.
Conflict of interest
This article does not have any conflict of interest.
Author’s contribution
SS and NA designed the study. SS and HA and IR conducted the experiments. RA and IR drafted the manuscript. WS, RA and SI analyzed the data. All authors critically reviewed the manuscript for intellectual content and gave final approval for final version.
REFERENCES