The Journal of Advances in Parasitology
Research Article
Molecular and Phylogenetic Status of Coenurus cerebralis Infecting Sheep from Dakahlia Province, Egypt
Ibrahim Abbas1*, Mohammed Elbeskawy2
1Parasitology Department, 2Internal, Infectious and fish diseases Department, Faculty of Veterinary Medicine, Mansoura University, Egypt - 35516.
Abstract | The limited genetic data as well as the economic and zoonotic importance of the genus Taenia, interestingly T. multiceps, from Egypt drew our attention to study the molecular characters and the phylogenetic position of this parasite species from sheep. Brains from 80 apparently healthy sheep slaughtered at Mansoura abattoir, Dakahlia province, Egypt, were incised and palpated for the presence of Coenurus cerebralis cysts. DNA was extracted from each cyst and amplified by PCR using two mitochondrial genes, the cytochrome oxidase (CO1) and the NADH dehydrogenase (ND1). Sequences were aligned with those published on GenBank and subjected to the phylogenetic analysis. C. cerebralis cysts were found in 3 (3.7%) out of the examined sheep and the three isolates were identical in their sequences. There were 99% identity between our isolates and those from Turkish (KT217594) and Iranian (JQ710587) sheep at the level of CO1 gene which confirmed by the phylogenetic analysis. Both CO1 and ND1 phenograms evidenced the existence of genetic variants within T. multiceps from variable hosts within different geographical regions. Our results supposed the conspecificity between the sheep and goat isolates, and the potential existence of a quasi-species in T. multiceps isolates from cattle. Such data could help in the epidemiological studies and implementation of control regimens against this economically important parasite.
Keywords | Coenurus cerebralis, Molecular characterization, Phylogeny, Egypt, Sheep
Editor | Muhammad Imran Rashid, Department of Parasitology, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | May 17, 2016; Accepted | July 02, 2016; Published | August 15, 2016
*Correspondence | Ibrahim Abbas, Parasitology Department, Faculty of Veterinary Medicine, Mansoura University, Egypt; Email: [email protected]
Citation | Abbas I, Elbeskawy M (2016). Molecular and phylogenetic status of Coenurus cerebralis infecting sheep from Dakahlia province, Egypt. J. Adv. Parasitol. 3(4): 117-124.
DOI | http://dx.doi.org/10.14737/journal.jap/2016/3.4.117.124
ISSN | 2311-4096
Copyright © 2016 Abbas and Elbeskawy. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Coenurus cerebralis is a transparent wall cyst containing many protoscolices, and represents the metacestode of the tape worm ‘’Taenia multiceps’’. Domestic and wild canids constitute the predators, while a wide range of herbivores including sheep, goats, cattle, buffaloes, camels, yak and equines are the prey hosts, (Sharma and Chauhan, 2006). This parasite usually inhabits the central nervous system specially the left and right cerebral hemisphere (Desouky et al., 2011).
Gid or sturdy is a fatal disease resulting from infection of sheep and goats with T. multiceps, and characterized by nervous manifestations including circling, blindness, head deviation and high (5-24.61%) mortalities (Njau et al., 1988; Biyikoglu et al., 2001) with subsequent great economic losses (Bussell et al., 1997; Achenef et al., 1999). Various human cases were infected with C. cerebralis inhabiting the central nervous system (Benifla et al., 2007) and intraocular cavities, resulting in endophthalmitis and retinal detachment (Inechukwu and Onwukeme, 1991).
Coenurosis is quite common in sheep (Sharma and Chauhan, 2006). The prevalence of infection was reported in many countries like, Italy (0.35%) (Scala et al., 2007), Turkey (1.3%) (Akkaya and Vurusaner, 1998), Jordon (3%) (Abo-Shehada et al., 2002) and Iran (9.8%) (Oryan et al., 1994). In Egypt, Desouky et al. (2011) noted C. cerebralis infection in 100% of clinically coenurosis suspected sheep, while no cysts were recovered from the apparently healthy animals.
Mitochondrial DNA is widely used for molecular characterization of many parasites because of its high evolutionary rate more than the nuclear DNA (Gasser et al., 1999).
Molecular characterization of T. multiceps was firstly studied in sheep using cytochrome c subunit 1 (CO1) and NADH dehydrogenase 1 (ND1) mitochondrial genes by Varcasia et al. (2006) in Italy. This study identified three genetic variants which designated as Tm1, Tm2 and Tm3. Rostami et al. (2013) reported 25 haplotypes from Iranian sheep using 12S rRNA gene. Further studies were conducted on cattle in Turkey by Avcioglu et al. (2011) and in Italy by Varcasia et al. (2012a), and on goats in Iran by Oryan et al. (2010) and in United Arab Emirates by Varcasia et al. (2012b). As far as we know, no sequence knowledge is available from the Egyptian isolates of Coenurus cerebralis from sheep.
For this reason, this study was planned to illustrate the molecular characters of T. multiceps isolates from sheep in Egypt using CO1 and ND1 mitochondrial genes, and compare it to that derived from other hosts within different geographical regions.
Material and methods
Samples Collection
Three C. cerebralis cysts were obtained after palpation and incision of brains from 80 apparently healthy sheep slaughtered at Mansoura abattoir, Dakahlia Province, Egypt during one year study extending from March 2014 to February 2015. The viability of the recovered cysts was assessed after incubation at 37°C for 12 hours. Protoscolices were harvested, washed 3 times in a phosphate buffer saline and stored in ethanol 70% until used.
DNA Extraction and PCR Amplification
Genomic DNA was extracted from protoscolices of each cyst using the standard phenol/chloroform technique (Sambrook et al., 1989). The harvested DNA was stored at -20°C until used. The mitochondrial CO1 gene (nearly 390 bp) was amplified by PCR using the primer pair COI1 forward (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) and COI2 reverse (5′-AAAGAAAGAACATAATGAAAATG-3′), while ND1 gene (nearly 470 bp) was amplified with the primer pair NADH 1 forward (5′-AGATTCGTAAGGGGCCTAATA-3′) and NADH2 reverse (5′-ACCACTAACTAATTCACTTTC-3′) according to Bowles et al. (1992).
Polymerase Chain Reactions (PCR) were performed in a final volume of 25 μL containing 2 μL of template DNA, 1 μL (25 μM) of each primer, 0.7 μL (10 mM) dNTP mix, 3.5 μL of Taq buffer (10X), 0.35 μL Taq polymerase (5Prime Perfect TaqTm) under the following cycling conditions: initial denaturation (94°C, 5 min), followed by 30 cycles for each of denaturation (94°C, 30 s), primer annealing (55°C, 30 s) and (extension 72°C, 30 s), and the final extension step at 72°C, 5 min. PCR products were subjected to gel electrophoresis using 1% agarose gel stained with ethidium bromide. Purified PCR amplicons from gel were commercially sequenced. The obtained sequences were compared to those published in the Genbank using the BLAST system, and then they were aligned using the software Bioedit. Mega (vesrsion 6) software was used to calculate the pairwise nucleotide variation and Taijma neutral index values, and to construct the neighbor-joining phylogenetic tree. All data were exported to an Excel spread sheet (Microsoft Office Excel 2010®).
Results
Three (3.7%) out of 80 apparently healthy sheep slaughtered at Mansoura abattoir, Egypt, were found to be infected with C. cerebralis. Cysts were located in the left cerebral hemisphere. Successful PCR amplification and partial genes sequencing were carried out for all the isolates. Results of the blast search molecularly identified the revealed cysts as T. multiceps metacestodes using the partial sequences of both CO1 (379bp) and ND1 (455bp) mitochondrial genes. The three isolates had the same sequences either for CO1 or ND1 genes.
For the CO1 partial sequences, the percent identities between our isolates and those previously published in Genbank were 96-99%. The highest identity (99%) was noted with the isolate from Iranian (JQ710587) and Turkish (KT217594) sheep with double nucleotides substitution (G/A) at positions 48 and 66 bp, and 5 nucleotides substitution with EF393620 and JX535575 from Turkey and China, respectively. The lowest identity was recorded with the isolate (HM143884) from the Turkish cattle (16 nucleotides substitution) (Figure 1).
Ninety nine percent of identity (triple nucleotides substitution) were revealed from the alignment of the ND1 gene partial sequences of T. multiceps isolates in the present study with the reference sequences KC794811 (A12G, G93A and G447T) and KT253936 (A12G, G93A and A168G) from the Chinese and Iranian goats, respectively. Moreover, the Turkish cattle isolate (HM143887) showed 98% identity in comparison to our isolates with 8 nucleotides substitution (Figure 2).
Pairwise comparison between the CO1 sequences of our isolates and the other T. multiceps isolates showed differences ranging from 0.9 to 4.7%, while among all the isolates, it ranged from 0.0 to 4.7%. At the level of ND1
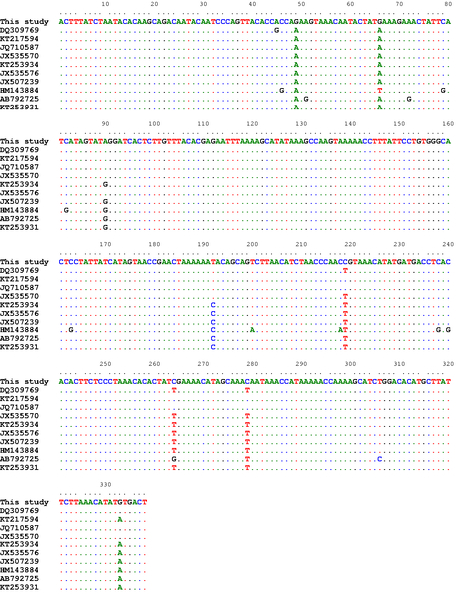
Figure 1: Alignment of the CO1 partial sequences
Alignment of the CO1 partial sequences (338bp) of our isolates with a number of T. multiceps isolates from variable host species within different geographical regions including sheep (DQ309769, KT217594, JQ710587, JX535570), goats (KT253934, JX535576, JX507239), cattle (HM143884) and dogs (AB792725, KT253931)

Figure 2: Alignment of the ND1 partial sequences
Alignment of the ND1 partial sequences (419bp) of our isolates with a number of T. multiceps isolates from variable host species within different geographical regions including sheep (DQ309770, DQ077820, KF233942, AJ239104, KT253935), goats (KT253936, KC794811), cattle (HM143887) and dogs (KT253932)
sequences, the values of the pairwise nucleotide variation were 1.0-2.2% by comparing our isolates with the other T. multiceps isolates and 0.0-3.1% among all the isolates.
Cluster analysis of the 31 studied T. multiceps CO1 partial sequences from a variety of hosts and geographical regions revealed 17 (0.54%) haplotypes with 23 segregation sites, while the haplotypes frequency of the ND1 gene was 0.63% (10 haplotype out of the 16 studied partial sequences) with 24 segregation sites. In addition, negative values of the Taijma D neutral index was reported for both genes (-0.985234 for CO1 and -1.320112 for ND1).

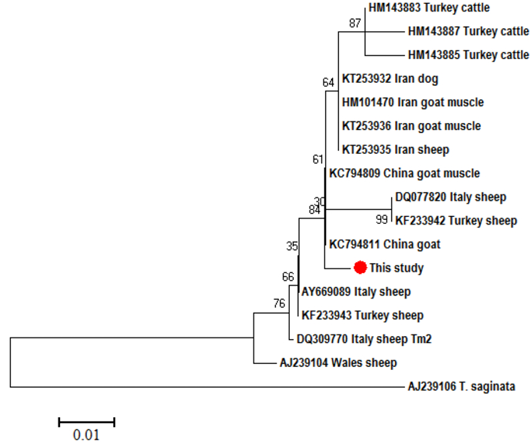
Figure 3: Neighbor-Joining phylogenetic tree
Neighbor-Joining phylogenetic tree of T. multiceps CO1 partial sequences from different hosts and geographical regions. T. saginata was used as an out group. Sequences were aligned by Bioedit and the tree was built using the software MEGA (version 6) with the Kimura 2 parameter mode. Scale bar indicates the proportion of sites changing along each branch
Using T. saginata as an out group, the phylogenetic analysis of T. multiceps CO1 partial sequences revealed 2 main clades, one of them consisted of our isolates along with that from Turkish (KT217594) and Iranian (JQ710587) sheep, while in the other clade, the isolates from sheep, goats, cattle and dogs were intermingled with each other’s (Figure 3). In the ND1 tree, also 2 main clades were formed, a common clade in which the sheep, goat, cattle and dogs isolates were intermixed with each other and separated from the other clade of sheep from Wales (AJ239104) (Figure 4).
Discussion
Coenurosis is a serious disease responsible for high economic losses in sheep industry, in addition to its zoonotic impact. C. cerebralis considered as the principal cause for nervous manifestations in sheep and goats from Egypt Desouky et al. (2011). Brain examination of 80 apparently healthy sheep slaughtered at Mansoura abattoir, Egypt, revealed the presence of C. cerebralis cysts in 3 (3.7%) cases. In Mansoura abattoir, no clinically diseased sheep were slaughtered. The uncontrolled home slaughter due to the week veterinary authorities, beside the high economic costs of sheep production due to high food prices, and the easily diagnosis of this disease by the farmers make them not to send their neurologically affected sheep for slaughtering in the official abattoirs fearing of condemnation. Although, neurological signs caused by C. cerebralis are common among sheep and goats, some animals remain normal and seemed to be healthy, and were diagnosed only after death (Sharma and Chauhan, 2006).
Neighbor-Joining phylogenetic tree of T. multiceps ND1 partial sequences from different hosts and geographical regions. T. saginata was used as an out group. Sequences were aligned by Bioedit and the tree was built using the software MEGA (version 6) with the Kimura 2 parameter mode. Scale bar indicates the proportion of sites changing along each branch
Previously in Egypt, C. cerebralis infection was estimated as 18.3% in a sheep flock from Suez Canal province (Anwar et al., 2013). Desouky et al. (2011) reported 100% prevalence in clinically diseased sheep from Cairo, although they did not found any cysts in the apparently healthy animals. Globally, different incidences were recorded, 44.4% from Tanzania (Miran et al., 2015), 100% and 2.7% from clinically diseased and apparently healthy sheep, respectively from Ethiopia (Achenef et al., 1999), 3% from Jordon (Abo-Shehada et al., 2002), 7.3% from Iraq (Karim, 1979), 18.7% from Iran (Tavassoli et al., 2012), 0.35% from Italy (Scala et al., 2007) and 15.5% from Turkey (Gicik et al., 2007). Incidence variation in the different geographical zones may be attributed to the varied geographical, sociological and ecological factors (Sharma and Chauhan, 2006).
Although, being an economically important parasite, no genetic data about C. cerebralis from Egypt are available as far as we know. The three revealed cysts in the present study were subjected to PCR amplification and sequencing using the mitochondrial genes CO1 (cytochrome oxidase subunit 1) and ND1 (NADH dehydrogenase 1) which have been used successfully for genetic studies on T. multiceps (Varcasia et al., 2006). For the first time, we molecularly confirmed the infection of sheep from Egypt with T. multiceps metacestodes.
Table 1: T. multiceps partial CO1 sequences originated from variable intermediate and definitive hosts within different geographical regions used in this study
|
Country |
Animal |
Tissue |
Accession number |
Reference |
|
Italy |
sheep |
brain |
DQ309767 DQ309768 DQ309769 |
Varcasia et al. (2006) |
|
FJ744755 |
unpublished |
|||
|
Turkey |
sheep |
brain |
EF393620 |
unpublished |
|
KT217580 KT217585 KT217588 KT217594 KT217595 |
unpublished |
|||
|
cattle |
HM143882 HM143884 HM143886 |
Avcioglu et al. (2011) |
||
|
Iran |
sheep |
brain |
KT253933 |
unpublished |
|
JQ710579 JQ710581 JQ710582 JQ710587 |
Rostami et al. (2013) |
|||
|
goat |
muscle |
KT253934 |
unpublished |
|
|
HM101469 |
Oryan et al. (2010) |
|||
|
CSF |
KP325489 |
|||
|
dog |
intestine |
KT253931 |
||
|
China |
sheep |
brain |
JX535570 JX535573 JX535575 |
unpublished |
|
goat |
JX535576 |
|||
|
JX507230 |
||||
|
muscle |
JX507239 |
|||
|
JX507238 |
||||
|
Mongolia |
dog |
intestine |
AB792725 |
unpublished |
|
Iran / T. saginata |
human |
intestine |
KJ910288 |
unpublished |
The alignment between our isolates and those of T. multiceps from different hosts and geographical regions at the level of both genes (CO1 and ND1) partial sequences, revealed a maximum identity (99%) with isolates from Turkish, Iranian, Italian and Chinese sheep and goats, while the minimum identity was reported with that from cattle.
The further molecular analysis of the studied T. multiceps partial sequences (Table 1 and 2) suggested the high genetic diversity within this parasite isolates. Considerably high haplotypes frequencies (0.54% for CO1 and 0.63% for ND1) and pairwise nucleotide variation values (up to 4.7% for CO1) were noted. Our results are coincided with that reported by Rostami et al. (2013) from the Iranian sheep (0.3-6.3%) using the CO1 gene. The high degree of nucleotide variation was observed particularly with the cattle isolates from Turkey. The expected high genetic diversity is attributed to the transmission of this parasite among a variety of hosts and the fact that during its life cycle, a single oncosphere give rise to a coenurus containing several hundreds of protoscolices and thus, one single mutant onchosphere can produce many genetically mutant identical worms which are capable of making new genetic variants (Rostami et al., 2013).
Table 2: T. multiceps partial ND1 sequences originated from variable intermediate and definitive hosts within different geographical regions used in this study
|
Country |
Animal |
Tissue |
Accession number |
Reference |
|
Italy |
sheep |
brain |
AY669089 DQ309770 DQ077820 |
Varcasia et al. (2006) |
|
Turkey |
sheep |
brain |
KF233943 KF233942 |
unpublished |
|
cattle |
HM143883 HM143887 HM143885 |
Avcioglu et al. (2011) |
||
|
Wales |
sheep |
brain |
AJ239104 |
Gasser et al. (1999) |
|
Iran |
sheep |
brain |
KT253935 |
unpublished |
|
goat |
muscle |
KT253936 |
unpublished |
|
|
HM101470 |
Oryan et al. (2010) |
|||
|
dog |
intestine |
KT253932 |
unpublished |
|
|
China |
goat |
brain |
KC794811 |
unpublished |
|
muscle |
KC794809 |
|||
|
Australia / T.saginata |
cattle |
muscle |
AJ239106 |
Gasser et al. (1999) |
Moreover, the negative values of the Taijma neutral index for both genes in the present study are indicators for the population expansion. Circulation of T. multiceps among different intermediate (sheep, goat, cattle, buffalo, horse and yak) and definitive hosts (dogs and red foxes), localization of its metacestodes in different tissues (brain, spinal cord, muscles and subcutaneous tissues), and the increased animal transportation between the export and import countries, could explain the worldwide distribution of this parasite species.
Referring to the phylogenetic analysis of the CO1 partial sequences, our isolates were distinguished as well as the sheep isolates from Turkey (KT217594) and Iran (JQ710587) in a separate clade, while all the other isolates were intermingled in another clade including sheep, goat, cattle and dog isolates. In the ND1 phylogenetic tree, our isolates were clustered nearer to the goat isolate (KC794811) from China in a clade consisted of different isolates from variable host species and distinct from the sheep isolate (AJ239104) from Wales. By looking to these data, both trees strengthen the evidences for the existence of genetic variants within T. multiceps from different geographical regions. In sheep, Varcasia et al. (2006) noted 3 genetic variants (Tm1, Tm2 and Tm3) from Italy using CO1, while, Rostami et al. (2013) reported 7 and 25 representative haplotypes using CO1 and 12S rRNA, respectively from Iran.
In the present study, the goat isolates, either originated from brain or muscles, were clustered within the sheep isolates, which indicates the absence of genetically variable strains in sheep and goats, like what have been previously said by Rostami et al. (2013), and opposite to Varcasia et al. (2012b) who suggested the existence of 2 different strains in sheep and goats. Akbari et al. (2015) stated that the cerebral and non-cerebral originated coenuri in goats after experimental infection by T. multiceps from dogs are identical morphologically and molecularly, although the cysts tend to be formed muscularly more than cerebrally.
Regarding the Turkish cattle isolates in the present study, clustering of these isolates in the phenogram with each other in a separate long branch within the main clade of sheep, goats and dogs` isolates as well as the high pairwise nucleotide variation (4.7%) proposed the potential existence of a quasi-species of T. multiceps originated from cattle. Avcioglu et al. (2011) and Varcasia et al. (2012b) suggested the presence of different strain of T. multiceps in cattle. Although, Varcasia et al. (2012a) noted the sheep Tm1 genetic variant in a cattle bull from Italy.
In conclusion, providing a precise picture about the existence of different strains within T. multiceps requires more molecular studies with efficient number of specimens from different predator and prey hosts specially cattle. Such molecular data may help in implementation of control regimens which considered as a difficult task and unsatisfactory to date.
Acknowledgments
We wish to thank the veterinarian staff at Mansoura Abattoir, Egypt, for their help in collecting the samples
Conflict of Interests
The authors declare that there is no conflict of interests.
Authors’ Contribution
Ibrahim Abbas carried out the laboratory work and wrote the manuscript, while Mohammed Elbeskawy collected the samples and helped in the laboratory work.
References