Advances in Animal and Veterinary Sciences
Research Article
Detection of Virulence invA Gene in the Salmonella Isolated from Fecal Samples of Poultry by Pcr Method
Mrunalini M. Pawade1*, Ashok V. Bhosale2, Prashant P. Mhase1, Mahesh B Kulkarni2, Shridhar D Budhe2
1*Department of Microbiology, KNP, College of Veterinary Science, Shirwal; 2Department of Microbiology, College of Veterinary Science, Udgir; India.
Abstract | The presence of Salmonella spp in collected faecal samples was assessed by performing the pre-enrichment and enrichment culture, followed by PCR assay. The primer was selected from the invA gene, specific for the detection of Salmonella spp. It was observed that 28% (10/35) of Salmonella were isolated by the conventional method. All Salmonella isolates thus obtained were subjected to PCR for the invA gene and all were found positive. In order to provide a more accurate profile of the prevalence of Salmonella spp in faecal samples, it was pertinent to use invA gene specific PCR method that could be considered as rapid technique for identification of Salmonella spp.
Keywords | Salmonella, InvA gene, Polymerase chain reaction (PCR), Faecal and Simmon citrate agar.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | July 05, 2017; Accepted | August 18, 2017; Published | November 23, 2017
*Correspondence | Mrunalini M Pawade, Department of Microbiology, KNP, College of Veterinary Science, Shirwal, India; Email: [email protected]
Citation | Pawade MM, Bhosale AV, Mhase PP, Kulkarni MB, Budhe SD (2017). Detection of virulence invA gene in the salmonella isolated from fecal samples of poultry by pcr method. Adv. Anim. Vet. Sci. 5(12): 520-522.
DOI | http://dx.doi.org/10.17582/journal.aavs/2017/5.12.520.522
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 Pawade et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Salmonella has been implicated as an important food-borne pathogen and thus has been the focus of many epidemiological studies in food animal populations. The majority of epidemiological studies of Salmonella still rely on conventional bacteriological culture methods to detect Salmonella in fecal samples (Davies et al., 2000; Funk et al., 2000). Culture-based methods for Salmonella are laborious and time-consuming, taking up to 7 days to complete, and are inefficient for epidemiological studies in populations with low prevalence of Salmonella, particularly because of the large number of samples that are typically required (Rostagno et al., 2005; Ward et al., 2005).Thus, a rapid, sensitive, and inexpensive method for the detection of Salmonella in fecal samples would enhance the efficiency of epidemiological studies of Salmonella. For accurate profiling of the prevalence of Salmonella spp, the use of invA gene specific PCR method is considered as an appropriate method. The polymerase chain reaction (PCR) offers a simple and reliable tool for the rapid detection of Salmonella spp.
Hence, in the present study, PCR was performed for specific detection of Salmonella spp. targeting the invA gene.
MATERIAL AND METHODS
Isolation and Identification of Salmonella Spp
In the present study, faecal samples of poultry (n=35) were collected and were processed for isolation of Salmonella spp by culturing into selenite F broth and incubated at 370C for 18 hours. After incubation, each broths were streaked on surface of Salmonella and Shigella agar plates (SS agar) and on Hecktoen Enteric agar (Andrew et al., 1979). Further these plates were incubated for 24 hrs at 370C. Presumptive black colonies like Salmonella were isolated and subjected to further biochemical characterization using Triple sugar iron agar (TSI agar) method and Simmons Citrate agar (Ateba, 2014).
Molecular Detection of the Isolates
DNA Extraction: DNA was extracted from the pure isolates by the boiling method, and for this approx. 1.5ml of the cultured broth was centrifuged at 10,000rpm for 5 minutes. The supernatant was discarded and the pellets were washed twice with sterile water. After this, 200µl of sterile water was added to the pellets, the pellets were vortexed to homogenize and boiled in a dry bath at 100°C for 10 minutes. This was followed by vortexing and centrifugation at 12,000rpm for 5 minutes. The supernatant containing the DNA was transfer to another tube. The purity of the extracted DNA was estimated using a Nanodrop spectrophotometer (Smith et al., 2015).
Primers set and PCR amplification: PCR Salmonellaspecific primers, S139 and S141 (Rahn et.al., 1992) have respectively the following nucleotide sequence based on the invA gene of Salmonella 5´ - GTG AAA TTA TCG CCA CGT TCG GGC AA - 3´and 5´ -TCA TCG CAC CGT CAA AGG AAC C -3´. Reaction with these primers were carried out in a 20μl amplification mixture consisting of 10μl of PCR Master mix (Qiagen), 20 pmol of each primer, 3μl of molecular grade water and 5μl of DNA were used in the reaction.
Amplification was conducted in Master-gradient Thermocycler (Eppendorf). The cycle conditions were as follow: An initial incubation at 940C for 60 sec. Followed by 35 cycles of denaturation at 94oC for 60 sec, annealing at 64oC for 30 sec and elongation at 72oC for 30 sec, followed by 7 min final extension period at 72oC.
Electrophoresis of PCR products: The amplified DNA products from Salmonella specific-PCR were analyzed with electrophoresis on 1.2% agarose w/v gels stained with ethidium bromide and visualized by UV illumination. A current of 120 V was applied to each gel. Eight μl of PCR product mixed with two μl of 6X loading dye were loaded on to agarose gel. A 100 bp DNA ladder (Himedia make-MBT049-50LN) was used as a marker for PCR products.
RESULTS AND DISCUSSION
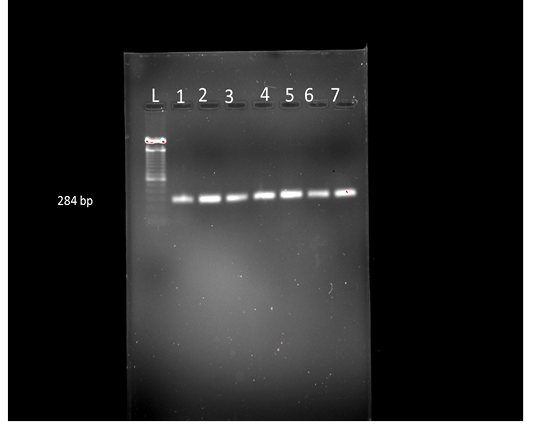
The present study supports the ability of these specific primer sets to confirm the isolates as Salmonella. From the 35 fecal samples, 10 isolates were found Salmonella positive as per conventional methods. On Triple sugar Iron agar, the salmonella showed alkaline reaction of the slant with red coloration of the medium, butt showed the acidic reaction with yellowing of the medium with slight gas production. All the isolates were able to utilize citrate as a sole carbon source and showed the change in colour from green to blue. All the 10 Salmonella positive isolates were screened by PCR for invA gene, and all of them resulted in 284 bp amplified fragment of invA gene (Karmi, 2013). Detection of invA in all the isolates proved that they were Salmonella species (Kocabiyik, 2006) (Figure 1). The invA gene contains sequences unique to the genus Salmonella and is considered the international standard for its identification (Malorny et al., 2003).
All Salmonella isolates, positive for the presence of invA gene which is responsible for invasion of cells, have the capacity to invade and survive in macrophages (Gole et al., 2013).The ability of Salmonella specific primers to detect Salmonella species rapidly and accurately is primarily due to the primer sequences that are selected from the gene invA. The invA gene codes for protein in inner membrane of bacteria, which is necessary for invasion to epithelial cells (Darwin and Miller, 1999). Rapid detection of Salmonella in poultry farms has an effective role in this study. PCR based methods with genus-specific primers belong to invA, according to its quick, specificity and sensitivity is a reliable technique for this proposes. In other hand this technique provides a tool, in confirmation of isolates as Salmonella. Although techniques which in recent years proposed for rapid and reliable detection and confirmation of Salmonella are very progress, but the PCR method which we use in our study, yet is a certain technique in identification and confirmation of Salmonella. The use of this method in most diagnostic and research laboratories is possible and through the molecular basis Salmonella identification techniques, this method is the simplest and a less expensive.
The amplification of the invA gene has been proposed as an international standard for genus of Salmonella detection (Malorny et al., 2003b). In the present study we used S138 and S141 primers for specific detection of Salmonella and the results of this study highlight the usefulness of the PCR for confirm detection of Salmonella spp from poultry faecal samples.
In conclusion, with attention to high level of Salmonella infections in poultry farms, it is necessary to consider control programs to prevention of economically loss resulting from mortality and spreading of infection.
ACKNOWLEDGEMENTS
The author wish to thank the faculty of Department of Microbiology, COVAS, Udgir for providing the facility to carry out the work.
CONFLICT OF INTEREST
The author declare that there is no conflict of interest.
Authors Contribution
M.M.Pawade: Administration of the whole research work, designing of methodology, standardizing the protocol and drafting of the final version of the article, A.V.Bhosale- Provision of the reagents and guidance of the work, P.P.Mhase-Drafting of the article, M.B.Kulkarni- Conducting the research work and S.D.Budhe- Collection of the samples, and drafting the article.
REFERENCES