Arginine Regulatory Mechanisms on Casein Synthesis in Dairy Cows
Arginine Regulatory Mechanisms on Casein Synthesis in Dairy Cows
Yifan Wang1,2, Liangyu Hu2, Numan Ullah2 and Mengzhi Wang2,3*
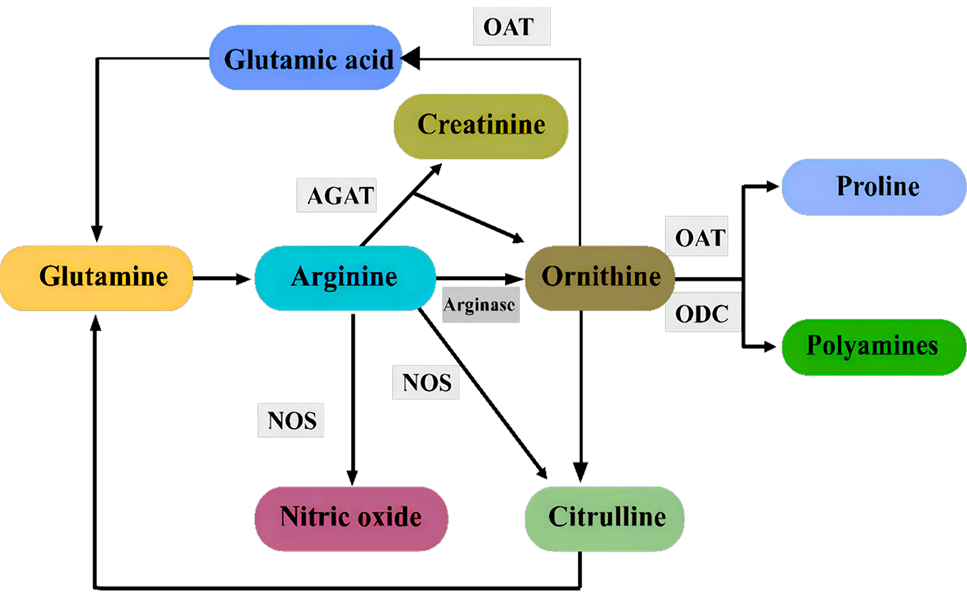
Arginine (Arg) metabolic pathways in mammary tissue. In the Arg/NO pathway, nitric oxide synthase (NOS) uses Arg to synthesize nitric oxide (NO) and citrulline. In the Arg/Orn pathway, Arg is converted to Ornithine (Orn) via arginase, and is further metabolized to proline or glutamine through transamination via ornithine aminotransferase (OAT) or to polyamines mainly through decarboxylation via ornithine decarboxylase (ODC). The third pathway of Arg metabolism is the arginine: Glycine amidino transferase (AGAT) in which Arg is used to synthesize Orn and creatinine.
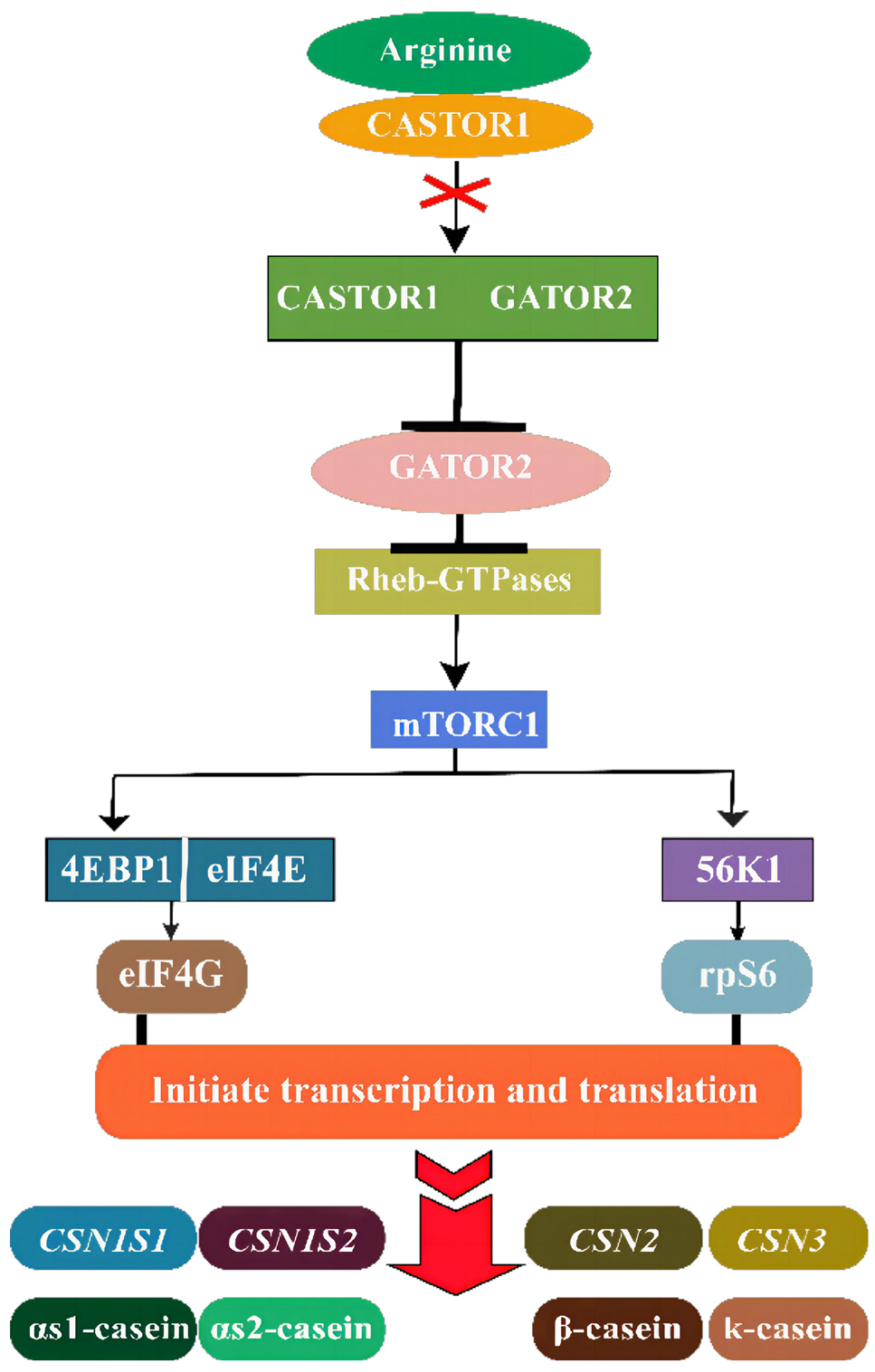
Arginine (Arg) -induced mechanistic target of rapamycin complex1 (mTORC1) signaling pathway in the context of casein synthesis. Arg disrupts the interaction of the cellular arginine sensor for mTORC1 (CASTOR) dimers with GATOR2 complex, allowing free GATOR2 to inhibit GATOR1 complex, and further activates mTORC1 through the active Rag GTPases to localize mTORC1 to lysosomes. Active mTORC1 activates transcription and translation of caseins through its interactions with eukaryotic initiation factor 4E-binding protein 1 (4EBP1), the eukaryotic initiation factor 4E (eIF4E), and ribosomal S6 kinase 1 (S6K1). Briefly, mTORC1 phosphorylates translation inhibitor 4EBP1, releasing it from eIF4E to free eIF4E to join the eukaryotic translation initiation factor 4G (eIF4G). mTORC1 phosphorylates and activates S6K1, which in turn stimulates the initiation of protein synthesis through activation of ribosomal protein S6 (rpS6).
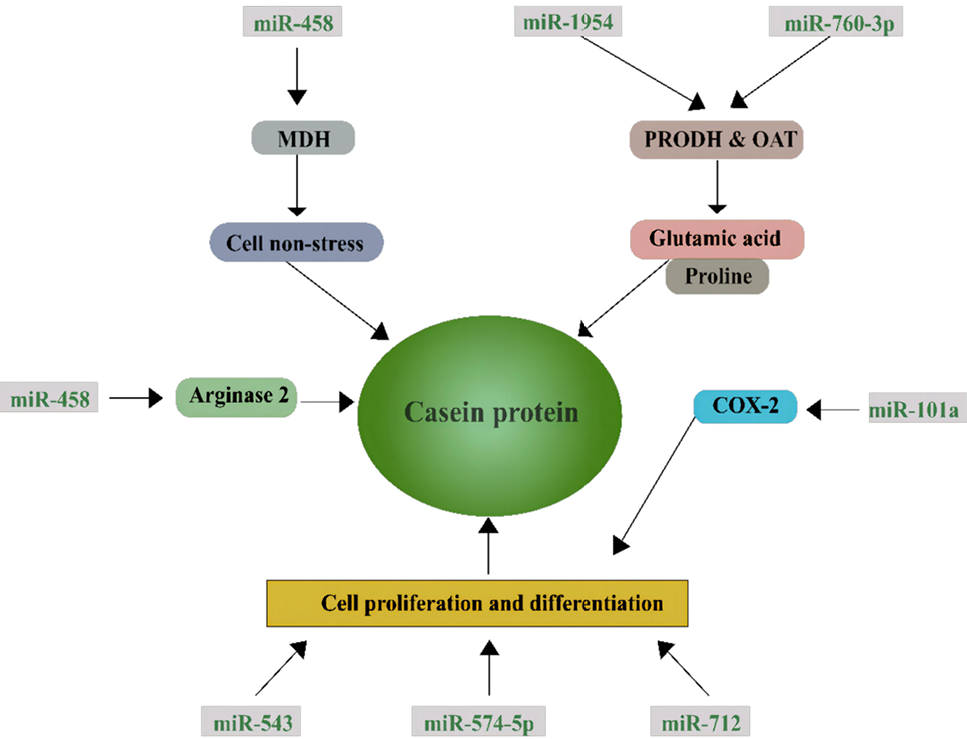
Mechanism of the regulation of 8 screened-miRNA by arginine (Arg) supplementation and effects on casein synthesis. Greater expression of miR-743a negatively regulates dehydrogenase (MDH) in response to supply of Arg leading to inhibition of MDH activity and, hence, alleviation of the oxidative stress status of mammary epithelial cells. miR-1954 and miR-760-3p are both predicted to target proline dehydrogenase (PRODH) and ornithine aminotransferase (OAT). Their induced expression with Arg supplementation suggested that Arg promotes cell proliferation and casein synthesis partly via inhibition on formation of glutamine and proline in the Arg/Orn pathway. miR-101a has been shown to regulate cell proliferation partly via alteration of the expression of cyclooxygenase-2 (Cox-2) that is critical for mammary gland development. Furthermore, miR-543, miR-574-5p and miR-712 target genes that are related to cell proliferation and differentiation, which suggests that Arg promotes cellular proliferation associated with those four miRNA candidates. The opposite response of miR-468 when the supply of Arg increases both in vitro and in vivo might be related to the regulation of arginase 2.