Complete Mitochondrial Genome of the Eurasian Oystercatcher Haematopus ostralegus and Comparative Genomic Analyses in Charadriiformes
Complete Mitochondrial Genome of the Eurasian Oystercatcher Haematopus ostralegus and Comparative Genomic Analyses in Charadriiformes
Chaochao Hu1,2, Xue Xu1, Wenjia Yao1, Wei Liu3, Deyun Tai1, Wan Chen4 and Qing Chang1*
The gene map of complete mitochondrial genome of the Eurasian Oystercatcher Haematopus ostralegus. Genes illustrated on the outside and inside of the main circle are encoded on the heavy (H) strand and the light (L) strand, respectively.
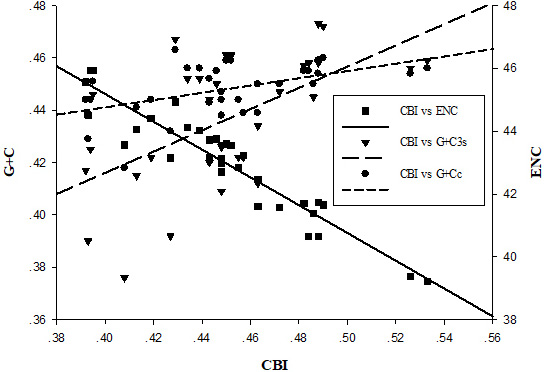
Evaluation of codon bias in the mitochondrial genomes of 32 species. ENC, effective number of codons; CBI, codon bias index; G + Cc, G + C content of all codon positions; G + C3s, G + C content of the third codon positions.
The molecular phylogenetic relationships of Haematopus ostralegus within the order Charadriiformes based on sequence of 13 PCGs and 2 ribosomal RNAs (12S rRNA and 16S rRNA) by using partitioned Bayesian analysis. The GenBank accession numbers for each species are shown in parentheses. Values on the branches represent bootstrap values/Bayesian posterior probabilities. “*” represented the nodes received the values of 1.00/100 unless otherwise labeled. The species which was sequenced in this study was underlined.