Detection, Serotyping and Antimicrobial Resistance Pattern of Salmonella from Chicken slaughter slabs in Jos, North-Central Nigeria
Detection, Serotyping and Antimicrobial Resistance Pattern of Salmonella from Chicken slaughter slabs in Jos, North-Central Nigeria
Idowu Fagbamila1*, Adaobi Okeke2, Micheal Dashen2, Patricia Lar2, Sati Ngulukun1, Benshak Audu1, David Ehizibolo1, Paul Ankeli1, Pam Luka1, Maryam Muhammad1
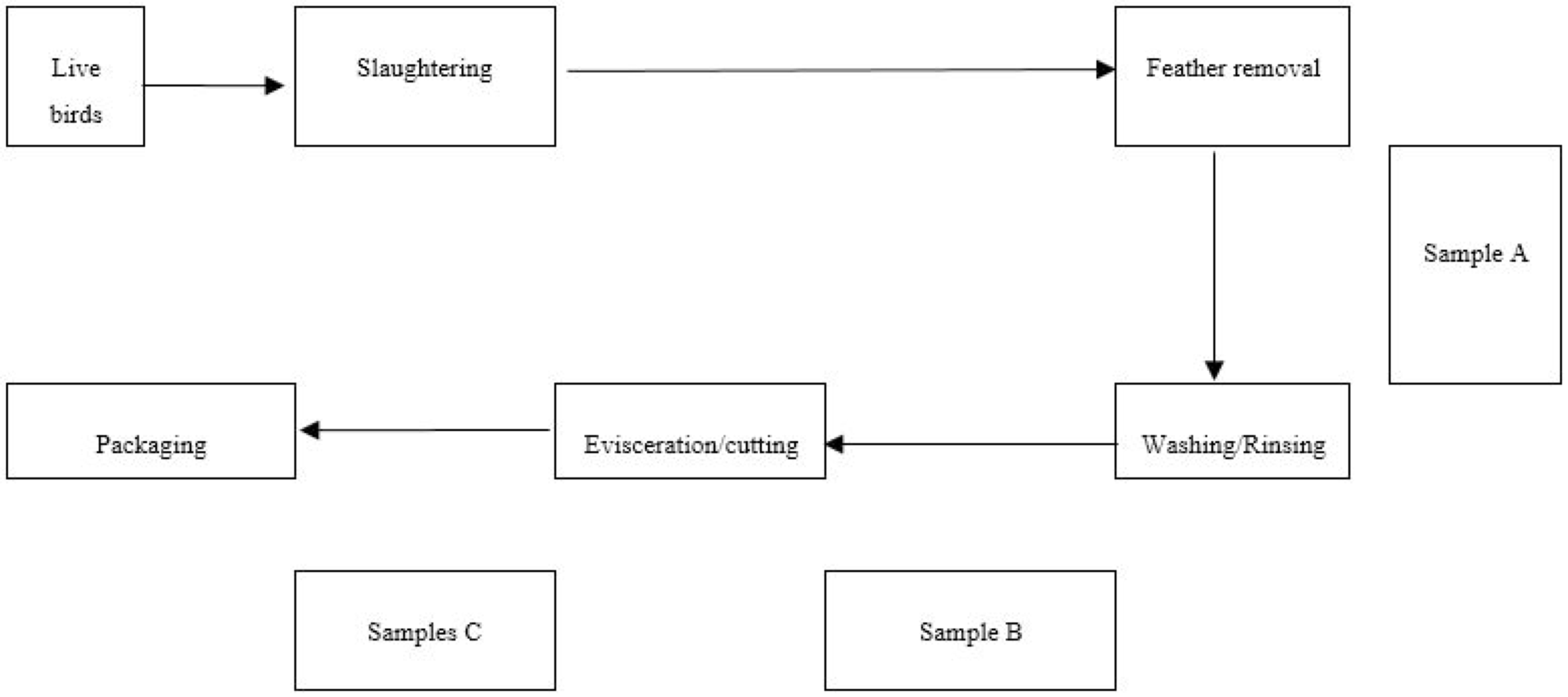
The different stages of processing of chicken at a slaughter slabs in a live bird market
Sample A: Defeathering (Warm) water; Sample B: Washing/Rinsing (Cold) water; Samples C: Intestinal contents and Processing surface (table swabs)
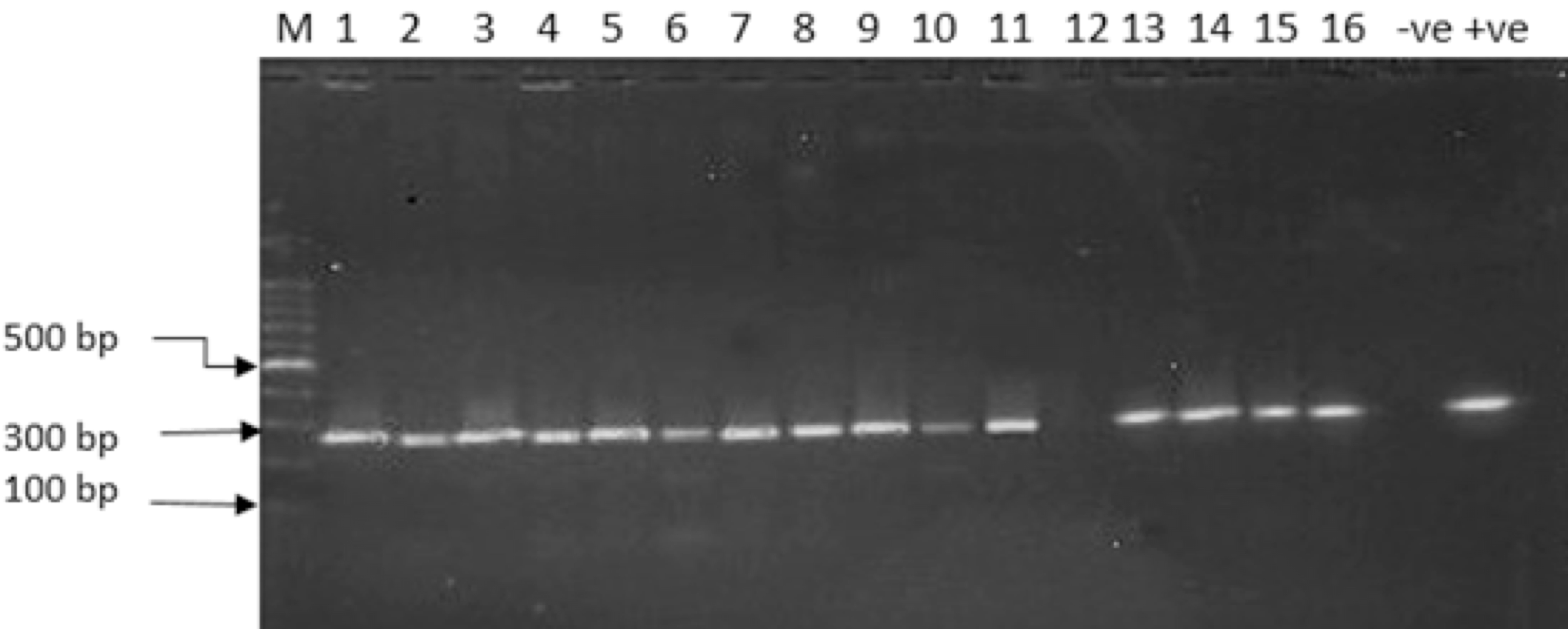
Agarose gel electrophoresis of amplified invA gene of 16 Salmonella isolates. Lane 1-16 represents Salmonella isolates; -ve represent negative control; +ve represent positive control and M represent the molecular weight marker, 100 bp ladder
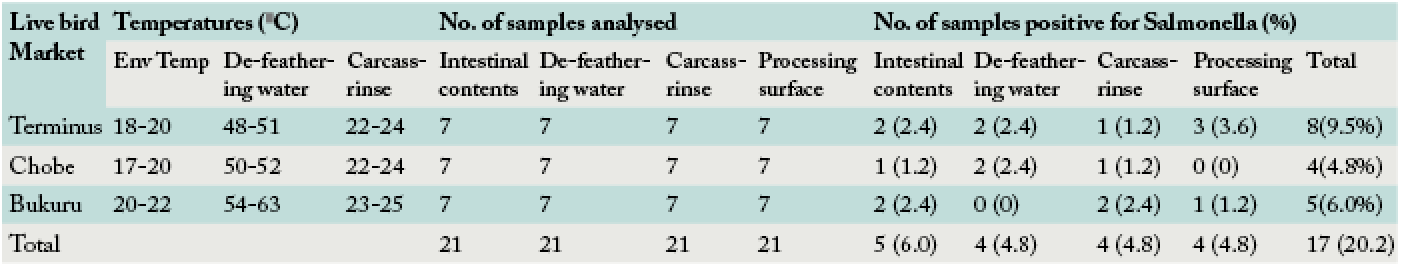
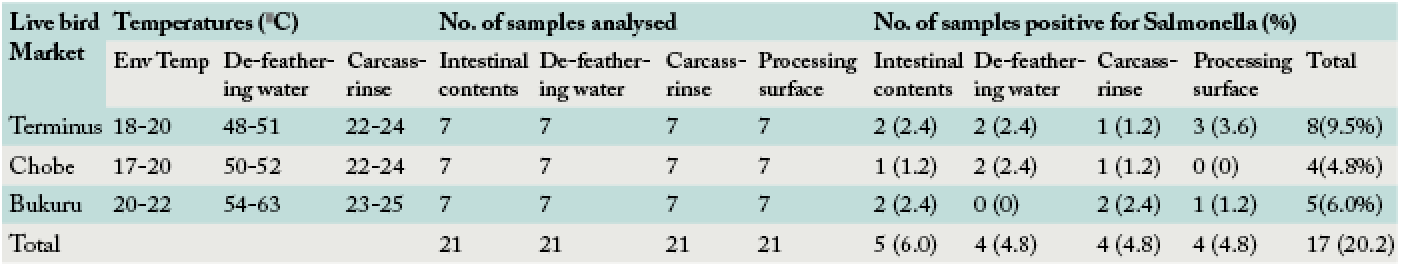
Isolation frequency of Salmonella from de-feathering water, carcass-rinse, intestine and processing surfaces at LBM in Jos, Plateau State, Nigeria
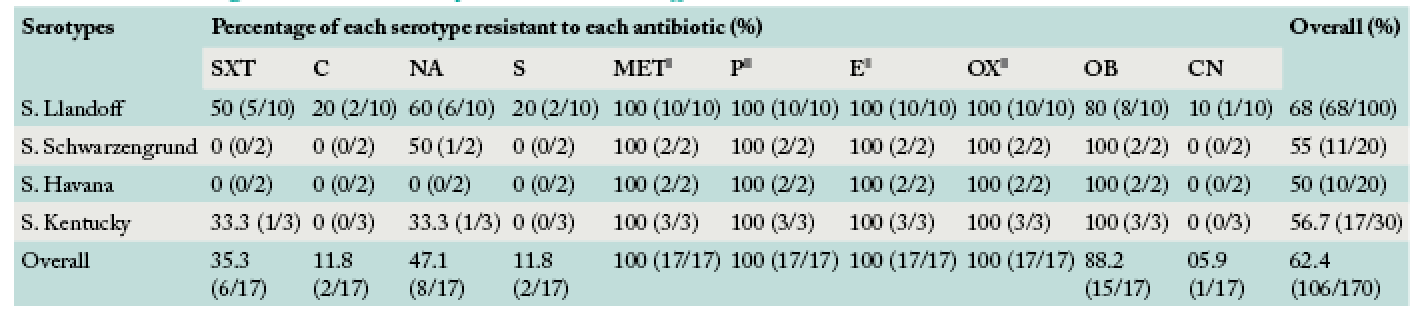
Percentage Salmonella serotypes resistant to different antibiotics
Isolation frequency of Salmonella from de-feathering water, carcass-rinse, intestine and processing surfaces at LBM in Jos, Plateau State, Nigeria