Genetic Diversity and Population Assignment of Arabian Horses
Mary A. Sargious1, Hanaa A. Ahmed1, Ragab M. El-Shawarby2, Hatem H. Bakery2, Sherif I. Ramadan3, E. Gus Cothran4 and Ayman Samir Farid5*
1Genome Research Unit, Animal Health Research Institute- ARC- 12618, Egypt.
2Department of Forensic Medicine and Toxicology, Faculty of Veterinary Medicine, Benha University, Moshtohor 13736, Egypt.
3Department of Animal Wealth Development, Faculty of Veterinary Medicine, Benha University, Moshtohor 13736, Egypt.
4Department of Veterinary Integrative Biosciences, College of Veterinary Medicine and Biomedical Sciences, Texas AandM University, College Station, Texas, 77843, USA.
5Department of Clinical Pathology, Faculty of Veterinary Medicine, Benha University, Moshtohor, Toukh 13736, Qalyubia, Egypt.
* Corresponding author: ayman.samir@fvtm.bu.edu.eg
Fig. 1.
Neighbor-joining phylogenetic tree of the three Arabian and the Dutch Warmblood horse populations based on16 microsatellite loci.
Fig. 2.
Structure clustering of the three Arabian and Dutch Warmblood horse populations obtained for K = 2.
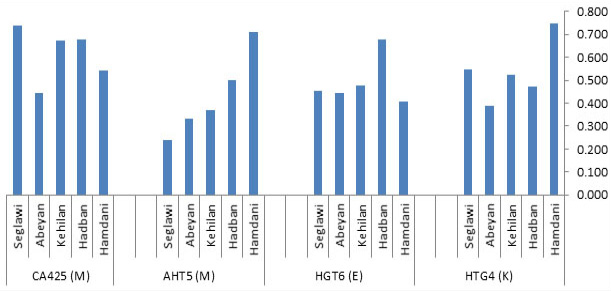
Fig. 3.
Allele frequencies’ distribution across the five basic horse strains “Al Khamsa”.
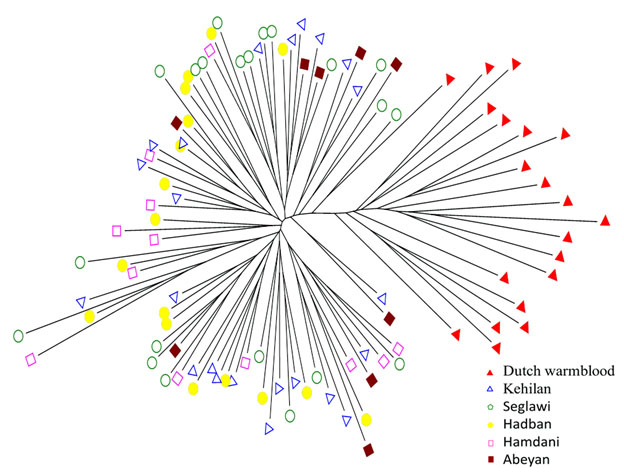
Fig. 4.
Individuals neighbor-joining phylogenetic tree across the five basic horse strains “Al Khamsa” in addition to Dutch Warmblood as an out group.
Fig. 5.
Structure clustering of the five basic horse strains “Al Khamsa” obtained for K = 2.