Genetic Diversity among Cotton Genotypes for Earliness, Yield and Fiber Quality Traits using Correlation, Principal Component and Cluster Analyses
Research Article
Genetic Diversity among Cotton Genotypes for Earliness, Yield and Fiber Quality Traits using Correlation, Principal Component and Cluster Analyses
Ghulam Sarwar1, Amna Nazir1*, Muhammad Rizwan1, Eram Shahzadi2 and Abid Mahmood3
1Cotton Research Station, Faisalabad (38000), Pakistan; 2Government College University, Faisalabad (38000), Pakistan; 3Directorate General of Agriculture Research, Faisalabad (38000), Pakistan.
Abstract | The upland cotton (G. hirsutum L.) is utmost natural fiber used in textile industry. Awareness about genetic diversity is very essential for fruitful genetic improvement approaches. Correlation, principal component analysis (PCA) and cluster analyses were performed to estimate genetic diversity in 25 cotton genotypes. Correlation analysis revealed that days to first square, flower and boll opening were positively associated with seed cotton yield. Staple length and CLCuV % were negatively associated with fiber fineness and yield respectively. In principal component analysis, four out of 14 principal components were selected with Eigen value ˃1. The four principal components contributed 79.9% towards variability. In PC-I, there was maximum positive factor loading of earliness traits, fiber fineness and negative factor loading of sympodia per plant and seed cotton yield. In PC-II, there was maximum positive factor loading of bolls per plant and GOT % while negative factor loading for CLCuV % and fiber length. A biplot between PC-I and PC-II showed that major contribution towards variability of fiber fineness and earliness traits among the studied genotypes. In cluster analysis, 25 genotypes were allocated in four clusters. Cluster-I was the largest by having 10 genotypes while clusters-III and IV each having six genotypes, and cluster-II was the smallest having three genotypes.
Received | October 06, 2020; Accepted | January 19, 2021; Published | March 08, 2021
*Correspondence | Amna Nazir, Cotton Research Station, Ayub Agricultural Research Institute, Faisalabad, Pakistan; Email: amnanazir.cri@gmail.com
Citation | Sarwar, G., A. Nazir, M. Rizwan, E. Shahzadi and A. Mahmood. 2021. Genetic diversity among cotton genotypes for earliness, yield and fiber quality traits using correlation, principal component and cluster analyses. Sarhad Journal of Agriculture, 37(1): 307-314.
DOI | http://dx.doi.org/10.17582/journal.sja/2021/37.1.307.314
Keywords | Correlation, Genetic divergence, Cluster analysis, Earliness, CLCuV, Seed cotton yield, Upland cotton
Introduction
Cotton is an important fiber crop of the world and a major source of our foreign exchange earnings. In cotton production, Pakistan ranks 4th in the whole world and it contributes 0.8% to GDP and 4.1% to value addition in agriculture (Anonymous, 2019). Its fiber industry comprises of 1815 textile and ginning units run by millions of the people which depends on cotton production (Khan, 2013). Due to its contribution in financial stability in more than 80 countries and multiple uses as lint and its by-products, it is called as “white gold” (Saeed et al., 2014; Dhivya et al., 2014). The upland cotton (G. hirsutum L.) has attracted valuable scientific interest of plant breeders due to its economic importance regarding yield and environmental suitability (Sun et al., 2019).
In Pakistan cotton production is low due to many biotic and abiotic factors. While considering the demand and importance of cotton crop, the development of high yielding and superior quality cotton genotypes is obligatory (Nazir et al., 2020). Therefore, the breeders are focusing to develop high yielding superior genotypes (Bakhtavar et al., 2015). Narrow genetic base and less diverse cotton genotypes are the main causes of yield decline (Rathinavel, 2017). So for broadening the genetic base of the cultivars, genetically distant genotypes should be used in developing high yielding cotton genotypes (Shakeel et al., 2015; Sun et al., 2019). To obtain superior genotypes it is necessary to properly exploit the available germplasm in the hybridization and introduction of new germplasm for creating ample genetic variation (Li et al., 2008).
Cotton seed yield being quantitative trait, depends on yield contributing traits and it could be increased by considering positive contribution of these yield components. A complete understanding about nature of crop, performance status and association of different agronomic traits with yield is necessary for researchers to eliminate the yield limiting factors. Nature and amount of genetic variance relies on different statistical methods used for assessment. Biometrical techniques i.e., principle component analysis (PCA), correlation analysis and cluster analyses have been repeatedly used identify the genetic diversity in different genotypes (Brown-Guedira et al., 2000). Estimation of genetic diversity through principal component analysis has made a way of recognition of phenotypic variability (Sun et al., 2019).The major advantage of PCA is that it arranges every single strain into only one set (Munir et al., 2020). Realizing the importance of genetic diversity, the present study was conducted in G. hirsutum genotypes to explore the variation among cotton germplasm for CLCuV, earliness, yield and fiber quality traits in context of recognition of model genotypes to be utilized in further breeding programs.
Materials and Methods
Breeding material and procedure
Twenty-five cotton elite lines viz. 5001, 5003, 5004, 5005, 5006, 5007, 5008, 5009, 5010, 5011, 5012, 5013, 5014, 5015, 5016, 5017, 5018, 5019, 5020, 5021, 5022, 5023, 5024, 5025 and check genotype FH-142 were evaluated during cropping season 2019-2020 at Cotton Research Station in Faisalabad, Punjab, Pakistan. The experimental material was sown on May 5th to find out genotypes having earliness, resistance against CLCuV and good seed cotton yield and fiber traits. The genotypes were planted in a randomized complete block design (RCBD) with three replications. Plot size for each entry was comprised of 6.09 × 3.04 m area with four rows and plants and rows spacing of 30 and 60 cm, respectively. All the recommended management practices including inputs were adopted.
Measurement of characters
Random selection of 10 plants was followed for measurement of traits. The data on days to first square, first flower and first boll opening was obtained by simply calculating days from sowing to emergence of first square, first flower and first opened boll respectively. Plant height (cm) was taken from main stem (base to top). Monopodia per plant and sympodia per plant were simply counted on the stem. Nodes to first fruiting branch were taken simply by counting the nodes from base to first fruiting branch appearance. The number of bolls were simply counted on whole plant. Seed cotton yield from experimental plot of each line was taken and converted to kg ha-1. Ginning was done with ginning machine having single roller and GOT % (ginning out turn) was calculated as:
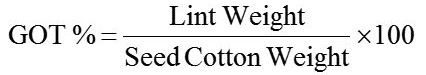
Fiber quality traits i.e., fiber length, fiber strength and fiber fineness for each plant was calculated by using USTER® HVI-1000. The CLCuV disease incidence (%) was taken according to disease scale (Table 1) given by Saeed et al., 2014 with formula:
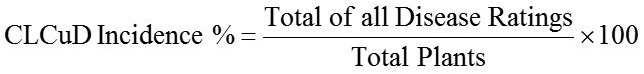
Statistical analysis
The two softwares i.e., Statistix 8.1 and Minitab-17 were used for basic statistics. Pearson correlation, cluster analysis and principal component analysis (PCA) were carried out according to Sneath and Sokal (1973).
Results and Discussion
Correlation studies
For studied traits, the basic statistics showed sufficient magnitude of variability among the 25 genotypes of cotton (Table 2 and Figure 1). Days to first square were significantly positively associated with days to first flower and boll opening and fiber fineness while significantly negatively associated with plant height, sympodia plant-1, seed cotton yield and fiber length (Table 3). Memon et al. (2016) findings showed contrary results of positive correlation of days to first square with yield. Days to first flower has significant positive association with days to first boll opening and fiber fineness, however, it revealed significant negative correlation with sympodia plant-1, yield and staple length. Days to first boll opening has significant positive association with fiber fineness and negative correlation with sympodia per plant, yield and staple length. Plant height showed significant positive association with sympodia per plant and yield but significant negative association with CLCuV %. Nikhil et al. (2018) also confirmed positive association of plant height with sympodia per plant. Farooq et al. (2014) also confirmed positive genotypic correlation of plant height with seed cotton yield and contrary findings for yield and earliness traits. Sympodia per plant has significant positive association with yield and staple length while revealed significant negative association with CLCuV % and fiber fineness. Salahuddin et al. (2010) also confirmed that sympodia per plant and yield were positively associated. Bolls per plant has significant negative association with CLCuV %. Yield is significantly positively associated with staple length and significantly negatively associated with CLCuV % and fiber fineness. Jarwar et al. (2019) also confirmed positive association of yield and staple length. Staple length has significant negative association with fiber fineness. Saeed et al. (2014) confirmed negative association of sympodia per plant with CLCuV %, fiber length and fiber fineness while positive correlation of days to first square with days to first flower, sympodia per plant with plant height and fiber length.
Principal component and cluster analyses
The variance is divided into its components for maintenance and exploitation of genetic diversity. Principal component analysis (PCA) is a useful technique to explore genotypes for successful breeding strategies (Akhter et al., 2009; Nazir et al., 2013). In this study, four out of 14 principal components were selected with Eigen value ˃ 1 (Table 4 and Figure 4). The contribution of these PCs towards total
Table 1: Rating scale for CLCuV disease symptoms.
|
Symptoms |
Disease ratings |
Disease incidence (%) |
Disease reaction |
|
No CLCuD symptoms |
0 |
0 |
Immune |
|
Only secondary and tertiary veins are swollen |
1 |
0.1-10 |
Highly Tolerant |
|
Primary veins, secondary and tertiary veins are swollen |
2 |
10-3 0 |
Tolerant |
|
Vein swelling, leaf curl or enation or both |
3 |
30-50 |
Susceptible |
|
Stunting alone with vein swollen, leaf curl/enation |
4 |
>50 |
Highly Susceptible |
Table 2: Basic Statistics for different Traits of cotton genotypes.
|
Traits |
Minimum |
Maximum |
Mean |
S.E. Mean |
Std. deviation |
|
Days to 1st square |
40 |
56 |
45.60 |
0.97 |
4.89 |
|
Days to 1st flower |
59 |
76 |
65.04 |
0.99 |
4.95 |
|
Days to 1st boll opening |
87 |
122 |
102.24 |
2.32 |
11.61 |
|
Plant height ( cm) |
102 |
184 |
131.52 |
4.08 |
20.44 |
|
Monopodia per plant |
2.0 |
6.0 |
3.68 |
0.18 |
0.94 |
|
Sympodia per plant |
18 |
32 |
25.60 |
0.80 |
4.04 |
|
Nodes to 1st fruiting branch |
4.0 |
8.0 |
5.96 |
0.20 |
1.01 |
|
Bolls per plant |
30 |
67 |
47.64 |
1.91 |
9.56 |
|
Yield kg/ha |
3944 |
6661 |
5283.6 |
177.24 |
886.19 |
|
CLCuv % |
12.5 |
32.3 |
21.96 |
1.06 |
5.33 |
|
GOT % |
39 |
45 |
41.96 |
0.31 |
1.56 |
|
Fibre length (mm) |
25.03 |
29.88 |
27.42 |
0.29 |
1.48 |
|
Fibre fineness (µg/inch) |
3.94 |
5.17 |
4.57 |
0.07 |
0.39 |
|
Fibre Strength (g/tex) |
28.18 |
34.71 |
31.82 |
0.29 |
1.45 |
Table 3: Correlation analysis (Pearson).
|
DFS |
DFF |
DBO |
PH |
SPP |
NFFB |
BPP |
Yield |
CL CuV |
GOT |
SL |
FF |
|
|
DFF |
0.91** |
|||||||||||
|
DBO |
0.93** |
0.88** |
||||||||||
|
PH |
-0.47* |
-0.2923 |
-0.3300 |
|||||||||
|
MPP |
0.2052 |
0.2698 |
0.2198 |
-0.1118 |
||||||||
|
SPP |
-0.81** |
-0.67** |
-0.75** |
0.72** |
||||||||
|
NFFB |
-0.1284 |
-0.1729 |
-0.1645 |
-0.0229 |
0.0566 |
|||||||
|
BPP |
-0.0423 |
0.1049 |
-0.0727 |
0.3564 |
0.2299 |
-0.35 |
||||||
|
Yield |
-0.90** |
-0.75** |
-0.84** |
0.68** |
0.91** |
0.08 |
0.18 |
|||||
|
CLCuV |
0.3552 |
0.3470 |
0.3624 |
-0.62** |
-0.63** |
0.06 |
-0.39* |
-0.57* |
||||
|
GOT |
0.0358 |
0.2041 |
0.0349 |
0.3231 |
0.1487 |
-0.14 |
0.04 |
0.1868 |
-0.19 |
|||
|
SL |
-0.71** |
-0.75** |
-0.79** |
0.0321 |
0.417* |
0.22 |
0.111 |
0.520* |
-0.11 |
-0.22 |
||
|
FF |
0.75** |
0.84** |
0.84** |
-0.0746 |
-0.51* |
-0.17 |
-0.08 |
-0.58* |
0.31 |
0.32 |
-0.87** |
|
|
FS |
-0.1345 |
-0.0836 |
-0.0896 |
0.2748 |
0.2186 |
-0.00 |
-0.14 |
0.2259 |
-0.24 |
0.03 |
0.002 |
-0.06 |
DFF: Days to first square; DFF: Days to first flower; DBO: Days to first boll open; PH: Plant height; MPP: Monopodia per plant; SPP: Sympodia per plant; NFFB: Nodes to first fruiting branch; BPP: Bolls per plant; CLCuV: Cotton leaf curl virus; GOT: Ginning out turn; SL: Staple length; FF: Fiber fineness; FS: Fiber strength.
variability was 79.9% which indicates that valuable information in first four components. The PC-I contributed maximum (44.7%), followed by PC-II (17%), PC-III (10.9 %) and PC-IV (7.3%). The present results were in accordance with the findings of Saeed et al. (2014), Latif et al. (2015), Kaleri et al. (2015), Shah et al. (2018) and Kumari and Gunasikaran (2019). The traits i.e., days to first square, flower, boll openingand fiber fineness displayed significant positive factor loadings in PC-I, while seed cotton yield and sympodia per plant had maximum negative loadings. Saeed et al. (2014) and Isong et al. (2017) also confirmed maximum contribution of sympodia per plant and yield in PC-I. Jarwar et al. (2017) also confirmed maximum contribution of yield in PC-I while contrary findings were reported by Shakeel et al. (2015). Maximum value of positive loadings on PC-II was observed in plant height, bolls per plant and GOT % whereas maximum negative factor loadings were revealed by CLCuV % and fiber length. These results were in conformity with the findings of Nazir et al. (2013) and Farooq et al. (2017) who reported that PC-II have maximum positive loadings for bolls per plant and GOT %. In PC-III, there was maximum positive factor loading for nodes to first fruiting branch and fiber strength, while negative factor loading for monopodia per plant and bolls per plant. Farooq et al. (2015) and Shakeel at al. (2018) also confirmed similar findings. In PC-IV, there was maximum positive factor loading for GOT %, monopodia per plant and CLCuV % while negative loading for bolls per plant. Latif et al. (2015) also mentioned positive factor loadings for GOT % and monopodia per plant while negative factor lodgings for CLCuV. The PCA expressed the magnitude of variability among the traits and this information may be exploited in further breeding programs for improvement in yield contributing traits (Nazir et al., 2013).
As depicted in PC biplot, the genotypes and variables were placed on the plot as vectors (Figure 2). The respective variable distances from PC-I and PC-II elaborated the contribution of different variables towards variability. Saeed et al. (2014) also studied main contribution of first two components in different cotton strains. Maximum contribution towards variability was shown by earliness related traits and fiber fineness in biplot. The biplot also showed the degree of correlation amongst parameters. Sympodia per plant and yield showed positive correlation with each other while negative correlation with CLCuV. While fiber fineness, days to first flower, days to first square and days to first boll opening were correlated with each other. As illustrated in biplot and score plot (Figures 2, 3), the genotypes 5006, 5012, 5003 and 5024 have good potential for fiber fineness, days to first flower, days to first square and days to first boll opening. While genotypes 5016, 5011 and 5008 may be selected for yield and sympodia per plant. The genotypes 5005. 5023 and FH-142 have good potential for staple length. The information acquired from Principle component analysis may be exploited to obtain valuable results.
Cluster Analysis is the assignment of a set of observations into subsets called clusters based on the similarity of the observations in the same cluster. A total of 25 genotypes were grouped in four groups. The cluster I, being the largest, comprised of 10 genotypes followed by cluster II, cluster III and cluster IV comprising 3, 6 and 6 genotypes, respectively (Table 6). As illustrated in Table 5, cluster I showed maximum values for earliness traits, while yield contributing
traits were maximum in cluster III. Cluster IV showed maximum values for fiber traits. Comparable results were observed by Shabbir et al. (2016), Farooq et al. (2017), Shakeel et al. (2018) and Jarwar et al. (2019), demonstrating prime values for yield contributing and fiber traits in the clusters. The dendrogram also showed the grouping of genotypes in clusters and sub-clusters (Figure 5). Based on cluster analysis the genotypes in cluster-I may be utilized for incorporation of earliness traits. The clusters-III and IV may be further exploited in breeding programs for the development of high yielding cotton genotypes with desirable fiber quality traits.
Table 4: Principal component analysis of different traits of cotton genotypes.
|
Variable |
PC I |
PC II |
PC III |
PC IV |
|
Eigen value |
6.2639 |
2.3820 |
1.5217 |
1.0174 |
|
% of total variance |
44.7 |
17.0 |
10.9 |
7.3 |
|
Cumulative variance % |
44.7 |
61.8 |
72.6 |
79.9 |
|
Factors loading by various characters |
||||
|
Days to 1st square |
0.379 |
0.055 |
0.065 |
-0.202 |
|
Days to 1st flower |
0.359 |
0.191 |
0.041 |
-0.098 |
|
Days to 1st boll opening |
0.375 |
0.115 |
0.105 |
-0.149 |
|
Plant height ( cm) |
-0.218 |
0.452 |
0.104 |
0.041 |
|
Monopodia per plant |
0.143 |
0.084 |
-0.514 |
0.389 |
|
Sympodia per plant |
-0.351 |
0.217 |
0.050 |
0.055 |
|
Nodes to 1st fruiting branch |
-0.068 |
-0.282 |
0.443 |
0.083 |
|
Bolls per plant |
-0.062 |
0.324 |
-0.469 |
-0.471 |
|
Yield kg/ha |
-0.369 |
0.165 |
0.021 |
0.169 |
|
CLCuv % |
0.225 |
-0.362 |
-0.081 |
0.340 |
|
GOT % |
0.010 |
0.397 |
0.120 |
0.550 |
|
Fibre length (mm) |
-0.294 |
-0.303 |
-0.200 |
-0.040 |
|
Fibre fineness (µg/inch) |
0.328 |
0.279 |
0.125 |
0.203 |
|
Fibre Strength (g/tex) |
-0.083 |
0.137 |
0.461 |
-0.233 |
Table 5: Cluster analysis of earliness, CLCuV, quality and yield traits of cotton genotypes.
|
Traits |
Cluster 1 |
Cluster 2 |
Cluster 3 |
Cluster 4 |
|
Days to 1st square |
50.50 |
46.33 |
41.17 |
41.50 |
|
Days to 1st flower |
69.00 |
67.33 |
61.67 |
60.67 |
|
Days to 1st boll opening |
113.40 |
107.00 |
93.83 |
89.67 |
|
Plant height ( cm) |
118.70 |
133.67 |
156.33 |
127.00 |
|
Monopodia per plant |
3.90 |
3.67 |
3.50 |
3.50 |
|
Sympodia per plant |
21.90 |
24.67 |
30.50 |
27.33 |
|
Nodes to 1st fruiting branch |
5.70 |
6.33 |
5.50 |
6.67 |
|
Bolls per plant |
46.20 |
41.67 |
54.17 |
46.50 |
|
Yield kg/ha |
4321.60 |
5312.00 |
6363.83 |
5792.67 |
|
CLCuv % |
24.61 |
24.99 |
16.51 |
21.49 |
|
GOT % |
41.70 |
42.33 |
42.83 |
41.33 |
|
Fibre length (mm) |
26.42 |
27.10 |
27.78 |
28.90 |
|
Fibre fineness (µg/inch) |
4.83 |
4.87 |
4.42 |
4.17 |
|
Fibre Strength (g/tex) |
31.49 |
32.09 |
31.83 |
32.25 |
Table 6: Cluster membership of cotton genotypes.
|
Cluster 1 |
10 |
5001, 5007, 5012, 5013, 5014, 5017, 5019, 5021, 5024, 5025 |
|
Cluster 2 |
3 |
5003, 5006, 5009 |
|
Cluster 3 |
6 |
5005, 5008, 5015, 5018, 5020, 5026 |
|
Cluster 4 |
6 |
5004, 5010, 5011,5016, 5022, 5023 |
Conclusions and Recommendations
The strains in cluster-III and IV can be pooled to acquire desirable combinations of earliness, less CLCuV %, good seed cotton yield and fiber quality traits. Earliness traits affects the CLCuV %, seed cotton yield, and fiber quality and. Therefore, earliness traits may further be explored to improve the seed cotton yield and fiber quality traits.
Novelty Statement
This is a novel approach to determine genetic variability in different cotton genotypes to identify group of genotypes having desirable combination of traits. The given research finally pointed out genotypes with desired traits.
Author’s Contribution
Ghulam Sarwar: Supervised and conducted research.
Amna Nazir: conducted research.
Muhammad Rizwan: helped in analysis.
Eram Shahzadi: helped in manuscript writing.
Abid Mahmood: overall supervision.
Conflict of interest
The authors have declared no conflict of interest.
References
Akhter, A., M.J. Hasan, A.K. Paul, M.M. Mutlib and M.K. Hossain. 2009. Selection of parent for improvement of restorer line in rice (Oryza sativa L.). SAARC J. Agric., 7: 43-50.
Anonymous, 2019. Pakistan Economic Survey. Finance Division, Economic advisor’s wing, Islamabad, Pakistan.
Bakhtavar, M.A., I. Afzal, S.M.A. Basra, A.U.H. Ahmad and M.A. Noor. 2015. Physiological strategies to improve the performance of spring maize (Zea mays L.) planted under early and optimum sowing conditions. PLoS One, 10(4): 1-15. https://doi.org/10.1371/journal.pone.0124441
Brown-Guedira, G.L., J.A. Thompson, R.L. Nelson and M.L. Warburton. 2000. Evaluation of genetic diversity of soybean introductions and North American ancestors using RAPD and SSR markers. Crop. Sci., 40: 815-823. https://doi.org/10.2135/cropsci2000.403815x
Dhivya, R., P. Amalabalu, R. Pushpa and D. Kavithamani. 2014. Variability, heritability and genetic advance in upland cotton (Gossypium hirsutum L.). Afr. J. Plant Sci., 8(1): 1-5. https://doi.org/10.5897/AJPS2013.1099
Farooq, J., M. Anwar, M. Riaz, A. Farooq, A. Mahmood, M.T.H. Shahid, M.R. Shahid and F. Ilahi. 2014. Correlation and path coefficient analysis of earliness, fiber quality and yield contributing traits in cotton (Gossypium hirsutum L.). J. Anim. Plant Sci., 24(3): 781-790.
Farooq, J., M. Rizwan, I. Sharif, S. Saleem, S.M. Chohan and R.A. Kainth. 2017. Genetic diversity studies in some advanced lines of Gossypium hirsutum L. for yield and quality related attributes using cluster and principle component analysis. AAB Bioflux., 9(3): 111-118.
Farooq, J., M. Rizwan, M. Anwar, M. Riaz, K. Mahmood and V. Petrescu-Mag. 2015.Multivariate analysis for CLCuD and various morphological traits in some advanced lines of cotton Gossypium hirsutum L. Int. J. Bioflux Soc., 7(3): 241-247.
Isong, A., P.A. Balu and P. Ramakrishnan. 2017. Association and principal component analysis of yield and its components in cultivated cotton. Electron. J. Plant. Breed., 8(3): 857-864. https://doi.org/10.5958/0975-928X.2017.00140.5
Jarwar, A.H., X. Wang, M.S. Iqbal, Z. Sarfraz, L. Wang, M.A.Qifeng and F. Shuli. 2019. Genetic divergence on the basis of principal component, correlation and cluster analysis of yield and quality traits in cotton cultivars. Pak. J. Bot., 51(3): 1143-1148. https://doi.org/10.30848/PJB2019-3(38)
Kaleri, A.A., S.Y. Rajput, G.A. Kaleri and J.A. Marri. 2015. Analysis of Genetic diversity in genetically modified and non-modified cotton (Gossypium hirsutum L.) genotypes. IOSR. J. Agric. Vet. Sci., 8(12): 70-76.
Khan, N.U., 2013. Diallel analysis of cotton leaf curl virus (CLCuV) disease, earliness, yield and fiber traits under CLCuV infestation in upland cotton. Aust. J. Crop Sci., 7(12): 1955-1966.
Kumari, V.N. and P. Gunasekaran. 2019. Evaluation of Genetic Diversity in Cotton (Gossypium barbadense L.) Germplasm for Yield and Fibre Attributes by Principle Component Analysis. Int. J. Curr. Microbiol. App. Sci., 8(04): 2614-2621. https://doi.org/10.20546/ijcmas.2019.804.304
Latif, A., M. Bilal, S.B. Hussain and F. Ahmad. 2015. Estimation of genetic divergence, association, direct and indirect effects of yield with other attributes in cotton (Gossypium hirsutum L.) using biplot correlation and path coefficient analysis. Trop. Plant. Res., 2(2): 120-126.
Li, Z., X. Wang, Y. Zhang, G. Zhang, L. Wu, J. Chi and Z. Ma. 2008. Assessment of genetic diversity in glandless cotton germplasm resources by using agronomic traits and molecular markers. Front. Agric. China, 2: 245-252. https://doi.org/10.1007/s11703-008-0063-x
Memon, S., G.H. Jatoi, N.A. Qureshi, M.J. Memon and L.A. Bhutto. 2016. Correlation estimates for earliness and yield related traits in cotton (Gossyium hirsutum L). Sci. Int. (Lahore)., 24(4): 4099-4102.
Munir, S., M.K. Qureshi, A.N. Shahzad, I. Nawaz, M.S. Anjam, S. Rasul and M.A. Zulfiqar. 2020. Genetic dissection of interspecific and intraspecific hybrids of cotton for morpho-yield and fiber traits using multivariate analysis. Pak. J. Agric. Res., 33(1): 9-16. https://doi.org/10.17582/journal.pjar/2020/33.1.9.16
Nazir, A., J. Farooq, A. Mahmood, M. Shahid and M. Riaz. 2013. Estimation of genetic diversity for CLCuV, earliness and fiber quality traits using various statistical procedures in different crosses of Gossypium hirsutum L. Vestnik OrelG. A. U., 43(4): 2-9.
Nazir, I., H.Z. Mahmood and T.M. Khan. 2020. Genetic variations in upland cotton (Gossypium hirsutum L.) for yield and fiber quality traits. Hortic. Int. J., 4(3): 83-89. https://doi.org/10.15406/hij.2020.04.00163
Nikhil, P.G., J.M. Nidagundi and A.A. Hugar. 2018. Correlation and path analysis studies of yield and fibre quality traits in cotton (Gossypium hirsutum L.). J. Pharmacogn. Phytochem., 7(5): 2596-2599.
Rathinavel, K., 2017. Exploration of genetic diversity for qualitative traits among the extant upland cotton (Gossypium hirsutum L.) varieties and parental lines. Int. J. Curr. Microbiol. App. Sci., 6(8): 2407-2421. https://doi.org/10.20546/ijcmas.2017.608.285
Saeed, F., J. Farooq, A. Mahmood, T. Hussain, M. Riaz and S. Ahmad. 2014. Genetic diversity in upland cotton for cotton leaf curl virus disease, earliness and fiber quality. Pak. J. Agric. Res., 27(3): 226-236.
Salahuddin, S., S. Abro, A. Rehman and K. Iqbal. 2010. Correlation analysis of seed cotton yield with some quantitative traits in upland cotton (Gossypium hirsutum L.). Pak. J. Bot., 42(6): 3799-3805.
Shabbir, R.H., Q.A. Bashir, A. Shakeel, M.M. Khan, J. Farooq, S. Fiaz, B. Ijaz and M.A. Noor. 2016. Genetic divergence assessment in upland cotton (Gossypium hirsutum L.) using various statistical tools. J. Glob. Innov. Agric. Soc. Sci., 4(2): 62-69. https://doi.org/10.22194/JGIASS/4.2.744
Shah, A.S., J.S. Khan, K. Ullah and O.U. Sayal. 2018. Genetic diversity in cotton germplasm using multivariate analysis. Sarhad J. Agric., 34(1): 130-135. https://doi.org/10.17582/journal.sja/2018/34.1.130.135
Shakeel, A., I. Talib, M. Rahid, A. Saeed, K. Zia and M.F. Saleem. 2015. Genetic diversity among upland cotton genotypes for quality and yield related traits. Pak. J. Agric. Sci., 52(1): 73-77.
Shakeel, A., M.T. Azhar, I. Ali, Q.U. Ain, Z.U. Zia, W. Anum and A. Zafar. 2018. Genetic diversity for seed cotton yield parameters, protein and oil contents among various bt. cotton cultivars. Int. J. Biosci., 12(1): 242-251. https://doi.org/10.12692/ijb/12.1.242-251
Sneath, P.H.A. and R.R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. W. F. Free-Man and Co., San Francisco., pp. 573.
Sun. Z., W. Xingfen, L.I.U. Zhengwen, G.U. Qishen, Z. Yan, L.I. Zhikun, K.E. Huifeng, Y. Jun, W.U. Jinhua, W.U. Liqiang, Z. Guiyin and M.A. Zhiying. 2019. Evaluation of the genetic diversity of fibre quality traits in upland cotton (Gossypium hirsutum L.) inferred from phenotypic variations. J. Cot. Res., 2(22): 1-8. https://doi.org/10.1186/s42397-019-0041-2
To share on other social networks, click on any share button. What are these?








