Genetic Variability and Heritability in F4 Populations of Brassica napus L.
Research Article
Genetic Variability and Heritability in F4 Populations of Brassica napus L.
Tahir Khan, Raziuddin, Fakharuddin* and Muazam Jamal Razi
Department of Plant Breeding and Genetics, The University of Agriculture, Peshawar-Pakistan.
Abstract | An experiment was conducted to estimate genetic variability, heritability and genetic advance for important attributes in Brassica napus using 10 parental lines and their 21 F4 populations. Significant differences were observed among genotypes, parents, F4 populations and parents versus F4 populations for days to 50% flowering, days to maturity, plant height, primary branches plant-1 and main raceme length. Among parents, CA2, DH8, DH4, CA4 and DH6 performed better for different traits. Whereas among F4 populations, cross combinations CA5 × DH8, CA2 × DH7, CA2 × DH8, CA5 × DH7, CA4 × DH7 and CA4 × DH8 performed better. Plant height and main raceme length displayed moderate to high broad sense heritability and maximum genetic advance for most of the F4 populations. Current results suggested the effectiveness of selection in early generations for the improvement of these traits.
Received | June 25, 2015; Accepted | February 23, 2016; Published | June 14, 2016
*Correspondence | Fakharuddin, The University of Agriculture, Peshawar-Pakistan; E-mail: fakharpbg@gmail.com
Citation | Khan, T., Raziuddin, Fakharuddin, and M.J. Razi. 2016. Genetic variability and heritability in F4 populations of Brassica napus L. Sarhad Journal of Agriculture, 32(2): 96-103.
DOI | http://dx.doi.org/10.17582/journal.sja/2016/32.2.96.103
Keywords | Brassica napus (rapeseed), Genetic variability, Heritability, Genetic advance
Introduction
Pakistan is chronically deficient in the production of edible oils and this deficit is continuously increasing due to the rapid rise in population and living standard of people. The local production of edible oil from all crops is only sufficient to meet one third of the domestic consumption while remaining being met through heavy imports by spending highly earned foreign exchange (FBS, 2014). Rapeseed and mustards share more than 20% in the domestic production which is the second largest share after cottonseed (FBS, 2014).
Rapeseed (mustards) is the second largest oilseed crop in the world providing 13% of the world’s edible oil after soybean. During 2014, rapeseed/ mustard were globally grown on area of 36.5 million hectare with the total production of 72.7 million metric tons having average yield of 1991.0 kg ha-1 (FAO Statistics, 2014). European Union is the leading producer followed by Canada, China, India and France. In Pakistan, during 2013-14 rapeseed was grown on an area of 24.70 thousand hectare, which produced 232.0 thousand tons seed with an average yield of 901.3 kg ha-1 (FAO Statistics, 2014). Due to increasing demand for rapeseed oil for food and non-food uses, the production of hybrid cultivars with higher seed and oil yields has become increasingly important (Ahmad et al., 2011). Rapeseed oil is used as food, as well as for industrial purposes including biodiesel production (Sabaghnia et al., 2010). A number of efforts have been undertaken to improve the rapeseed-canola cultivars for quality and production. Several genotypes/varieties have been developed and tested in different parts of Pakistan (Mahmood et al., 2011; Ahmad et al., 2012) but these varieties are not sufficient to fulfil domestic requirements. Furthermore, area under cultivation of oilseed crops in the country is also decreasing with the passage of time showing the negligence of national policies. Non-availability of better-adapted genotypes is a reason for decreasing acreage of these crops (Khan et al., 2006). Oilseed brassica being well entrenched in the cropping system of Pakistan can potentially reduce consumption and production gap and decrease burden on exchequer (Syed et al., 1994).
Genetic variability, heritability as well as genetic gain in selection contribute to success of any crop improvement program (Khan et al., 2006). Genetic variability is the difference in plants on the basis of different characters. Plant breeders are trying to explore and induce diversity among the available genetic material. This helps in the selection of desirable line(s) and it also helps in developing a new cultivar which is adaptable to diverse agro-climatic conditions. Therefore, diversity of plant genetic resources is imperative for crop improvement (Jatoi et al., 2012).
Estimates of heritability help in predicting performance of genotypes in succeeding generations and thus provide a vital component of response to selection for successful breeding programs. Heritability is a key of transmissibility of traits and as such partition the total variance into genetic and environmental components (Falconer and Mackay, 1996; Marwede et al., 2004). Heritability can also help in determining the choice of breeding system. It plays a main role in bringing improvement in the crop plants. Genetic gain is the amount of increase in performance that is achieved through artificial genetic improvement. This term is usually used to refer to the increase after one generation has passed. Thus by estimating the genetic advance breeders can get clear idea that how much improvement is brought after passing one generation. Keeping in view the importance of rapeseed as an important oilseed crop an experiment was conducted to evaluate Brassica napus L. genotypes and their F4 populations with the objectives to estimate genetic variability and broad sense heritability for important traits.
Materials and Methods
To study genetic variability, heritability and genetic advance in parents and F4 populations of Brassica napus, an experiment was conducted during 2014-15 at The University of Agriculture, Peshawar - Pakistan.
Brassica napus germplasm comprised of ten parents (7 lines viz., DH-2, DH-3, DH-4, DH-5, DH-6, DH7 and 3 testers viz., CH-8, CA-2, CA-4 and CA-5) and their twenty-one F4 populations. All genotypes were planted in randomized complete block design with three replications. Each genotype was planted in five rows replication-1 with the row length of 5m. Row-to-Row and plant to plant distance was kept at 50 cm and 30 cm, respectively. All agronomic practices were performed uniformly for all genotypes.
Data were recorded on 10 randomly selected plants from each genotype at proper time for days to flowering, days to maturity, number of primary branches on main stem, plant height and main raceme length.
Statistical analysis
The data was statistically analysed according to the appropriate method recommended for randomized complete block design (Steel and Torrie, 1980). Mean separation was carried out following LSD test. Broad sense heritability for a particular trait was computed using parental and F4 populations of each cross combination using the modified version of the formula suggested by Mahmud and Kramer (1951):
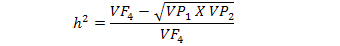
Where,
VF4 = Variance of F4 population for a specific trait.
VP1 and VP2 = Variances of parent 1 and parent 2 of a specific F4 population.
Genetic advance was computed by using following formula as suggested by Panse and Sukhatme (1965):
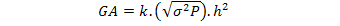
Where,
GA = Genetic advance, K = 1.76 for 10 % selection intensity, h2 = Heritability coefficient (broad sense), (√σ2P)= Phenotypic standard deviation
Results and Discussion
The results of the experiment based on important characters are discussed below:
Days to 50% flowering
In rapeseed, early flowering generally results in early maturity which helps plants to escape heat stress therefore early flowering is considered favourable.
Table 1: Mean squares of parents and F4 populations for various traits in Brassica napus
|
SOV |
Rep (df=2) |
Genotype (df=30) |
Parents (df=9) |
F4 (df=20) |
P vs F4 (df=1) |
Error |
CV% |
|
Days to 50% flowering |
7.3 |
9.4** |
14.0** |
5.1* |
51.7** |
2.7 |
1.51 |
|
Days to maturity |
17.8 |
56.5** |
106.1** |
26.2** |
215.5** |
12.5 |
2.14 |
|
Plant height |
83.9 |
444.2** |
160.7* |
330.0** |
5278** |
77.0 |
5.13 |
|
Number of primary branches |
0.3 |
2.7** |
3.6** |
2.2** |
4.1** |
0.8 |
0.8 |
|
Main raceme length |
36.1 |
183.7** |
166.7** |
185.4** |
302.8* |
63.3 |
12.22 |
* and **: Significant and highly significant at 0.05 and 0.01 level of probability, respectively
In the current experiment, genotypes parents and parents versus F4 showed highly significant difference (p≤0.01), whereas F4 populations showed significant differences at (p≤0.05) (Table 1). The results indicated the presence of sufficient variability in the tested genotypes. Similar findings have been reported earlier by Nazeer et al. (2003), Azam et al. (2013), Nasim et al. (2013) and Muhammad et al. (2014) who observed significant differences for days to flowering in Brassica napus genotypes.
On overall basis, mean values for days to 50% flowering varied from 106 to 112 with an average value of 109 days. Data for parental genotypes ranged from 106 to 112 days with the mean of 108 days where earliest flowering was observed in DH2 whereas flowering in CA2 took maximum days. Mean values of F4 population ranged from 108 to 112 days with mean of 110 days (Table 2). Earliest flowering was observed in CA2 × DH3, CA2 × DH7 and CA5 × DH3. Overall variances vary between 2.3 for the genotype CA2 to 31.6 for cross CA2 × DH4. Among parental genotypes, variance ranged between 2.3 for CA2 to 16.22 for DH4. In F4 populations, variance varied from 6 to 31.6 for CA2 × DH8 and CA2 × DH4, respectively (Table 3). Heritability estimations exhibited that majority of the populations had moderate to low heritability. Broad sense heritability in the F4 populations was lowest for CA2 × DH7 (0.31) and highest for CA5 × DH6 (0.63). Genetic advance was low for the cross CA4 × DH3 (1.4) while it was highest for the cross CA2 × DH4 (6.3). High heritability and genetic advance shows more genetic control and selection could be effective to bring improvement in days to flowering. Mahto and Haider (2002) and Chaghakaboodi et al. (2012) also reported similar results in rapeseed genotypes.
Days to maturity
Early maturity in rapeseed helps to escape heat stress and some late coming diseases therefore early maturing genotypes are preferred by the farmers. In present study, highly significant differences (P ≤ 0.01) were observed among genotypes, parents, F4 populations and parents versus F4 (Table 1). Zare and Sharafzadeh (2012) and Hasan et al. (2014) also reported significant differences for days to maturity among Brassica napus genotypes.
Mean data for days to maturity on overall basis showed that minimum days were taken by CA5 (153days) while the maximum days were taken by CA5 × DH8 (175days). Among parental genotypes, maturity fluctuated from 153 to 172 days for CA5 and CA2 respectively with a mean of 163 days. Among F4 populations, days taken to maturity were 162 to 175days for CA4 × DH8 and CA5 × DH8, respectively with the mean value of 166 days (Table 2).
On overall basis, variance for genotypes ranged between 2.17 to 19.10 for DH5 and CA2 × DH7, respectively. Among parents, smallest variance (2.16) was recorded for DH5 while the highest (13.2) was recorded for DH2 whereas, in F4 populations, the lowest variance (6.13) was displayed by CA4 × DH5, while the highest variance (19.1) was recorded for CA2 × DH7 (Table 3). Majority of the heritability estimates were moderate whereas few were low. The lowest value of heritability was recorded for CA5 × DH7 (0.32) while highest value was displayed by CA2 × DH8 (0.56). Lowest genetic advance was recorded for CA4 × DH (1.6) while the highest for CA2 × DH8 (4.1) (Table 3). Our results are partially in agreement with the earlier finding of Zare and Sharafzadeh (2012) and Rameeh (2014) who observed high heritability for days to maturity in Brassica napus.
Plant height (cm)
In brassica, medium sized plants are preferred because
Table 2: Means values of parents and F4 populations of important traits in Brassica napus
|
Genotypes |
Days to 50% flowering (days) |
Days to maturity (days) |
Plant height (cm) |
No. Primary Branches (nos) |
Main raceme length (cm) |
|
CA2 |
112 |
172 |
185 |
8.6 |
81 |
|
CA4 |
111 |
168 |
186 |
5.4 |
64 |
|
CA5 |
107 |
153 |
188 |
6.9 |
80 |
|
DH2 |
106 |
162 |
172 |
5.3 |
65 |
|
DH3 |
109 |
156 |
188 |
6.0 |
68 |
|
DH4 |
106 |
163 |
186 |
5.5 |
69 |
|
DH5 |
108 |
168 |
178 |
6.1 |
62 |
|
DH6 |
108 |
162 |
179 |
5.3 |
68 |
|
DH7 |
107 |
160 |
168 |
5.3 |
61 |
|
DH8 |
107 |
168 |
190 |
7.2 |
59 |
|
Parents mean |
108 |
163 |
182 |
6 |
68 |
|
CA2 × DH2 |
111 |
165 |
184 |
6.1 |
77 |
|
CA2 × DH3 |
108 |
164 |
175 |
4.2 |
75 |
|
CA2 × DH4 |
109 |
166 |
155 |
5.7 |
59 |
|
CA2 × DH5 |
109 |
166 |
143 |
4.6 |
50 |
|
CA2 × DH6 |
109 |
167 |
166 |
5.2 |
67 |
|
CA2 × DH7 |
108 |
166 |
165 |
5.2 |
74 |
|
CA2 × DH8 |
111 |
168 |
162 |
7.2 |
62 |
|
CA4 × DH2 |
109 |
170 |
162 |
5.7 |
55 |
|
CA4 × DH3 |
111 |
165 |
172 |
5.1 |
67 |
|
CA4 × DH4 |
112 |
172 |
183 |
7.7 |
71 |
|
CA4 × DH5 |
112 |
165 |
170 |
6.0 |
68 |
|
CA4 × DH6 |
112 |
166 |
164 |
6.1 |
63 |
|
CA4 × DH7 |
109 |
166 |
157 |
5.2 |
63 |
|
CA4 × DH8 |
109 |
162 |
169 |
5.3 |
59 |
|
CA5 × DH2 |
110 |
167 |
176 |
5.9 |
68 |
|
CA5 × DH3 |
108 |
166 |
157 |
4.9 |
63 |
|
CA5 × DH4 |
112 |
166 |
148 |
6.2 |
47 |
|
CA5 × DH5 |
109 |
165 |
163 |
5.7 |
69 |
|
CA5 × DH6 |
109 |
165 |
173 |
6.0 |
64 |
|
CA5 × DH7 |
110 |
163 |
176 |
4.8 |
64 |
|
CA5 × DH8 |
109 |
175 |
163 |
6.9 |
54 |
|
F4 mean |
110 |
166 |
166 |
6 |
64 |
|
Genotypes mean |
109 |
165.4 |
171 |
5.9 |
65 |
|
LSD(0.05) Parents |
0.58 |
1.26 |
3.13 |
0.31 |
2.84 |
|
LSD(0.05) F4`s |
0.85 |
1.83 |
4.53 |
0.45 |
4.11 |
|
LSD(0.05) Genotypes |
2.69 |
5.77 |
14.33 |
1.44 |
13.00 |
medium plants can withstand against lodging therefore in breeding experiments plant height is given due importance. In current study, highly significant differences (P ≤ 0.01) were observed for plant height among genotypes, parents versus F4 and F4 while significant differences (P ≤ 0.05) existed among parents (Table 1). Azam et al. (2013) and Muhammad et al. (2014) also reported significant differences for plant height in brassica genotypes.
On overall basis, plant height ranged from 143 to 190 cm for CA2 × DH5 and DH8, respectively. Plant height in the parents ranged between 168 to 190cm for DH7 and DH8 respectively with the average of 182 days
Table 3: Variances, heritability and genetic advance of days to 50% flowering, days to maturity and plant height in Brassica napus L
|
Genotypes |
Days to flowering (days) |
Days to maturity (days) |
Plant height (cm) |
||||||
|
Var. |
H2(bs) |
GA |
Var. |
h2(bs) |
GA |
Var. |
h2(bs) |
GA |
|
|
CA2 |
2.3 |
7.4 |
54.8 |
||||||
|
CA4 |
3.5 |
5.4 |
124.9 |
||||||
|
CA5 |
2.6 |
4.9 |
34.7 |
||||||
|
DH2 |
6.2 |
13.2 |
83.4 |
||||||
|
DH3 |
5.1 |
6.1 |
157.2 |
||||||
|
DH4 |
16.2 |
8.0 |
209.8 |
||||||
|
DH5 |
3.6 |
2.2 |
116.9 |
||||||
|
DH6 |
3.9 |
6.0 |
195.7 |
||||||
|
DH7 |
3.4 |
8.6 |
159.9 |
||||||
|
DH8 |
4.2 |
8.1 |
233.8 |
||||||
|
CA2 × DH2 |
19.2 |
0.59 |
4.5 |
17.1 |
0.41 |
3.0 |
123.6 |
0.44 |
11.5 |
|
CA2 × DH3 |
6.1 |
0.34 |
1.5 |
10.9 |
0.36 |
2.1 |
447.2 |
0.60 |
12.8 |
|
CA2 × DH4 |
31.6 |
0.63 |
6.3 |
12.1 |
0.39 |
2.4 |
342.1 |
0.53 |
20.6 |
|
CA2 × DH5 |
7.2 |
0.53 |
2.5 |
11.1 |
0.55 |
3.2 |
213.1 |
0.57 |
13.6 |
|
CA2 × DH6 |
24.6 |
0.58 |
5.1 |
10.1 |
0.37 |
2.0 |
294.2 |
0.55 |
17.7 |
|
CA2 × DH7 |
10.3 |
0.41 |
2.3 |
19.1 |
0.50 |
3.9 |
294.8 |
0.59 |
12.4 |
|
CA2 × DH8 |
6.0 |
0.47 |
2.0 |
17.2 |
0.56 |
4.1 |
659.2 |
0.55 |
21.3 |
|
CA4 × DH2 |
8.3 |
0.44 |
2.2 |
14.8 |
0.42 |
2.8 |
359.6 |
0.56 |
14.5 |
|
CA4 × DH3 |
6.4 |
0.31 |
1.4 |
9.9 |
0.39 |
2.2 |
253.3 |
0.43 |
8.6 |
|
CA4 × DH4 |
16.4 |
0.33 |
2.4 |
14.1 |
0.41 |
2.7 |
528.2 |
0.45 |
13.4 |
|
CA4 × DH5 |
6.9 |
0.47 |
2.2 |
6.1 |
0.38 |
1.6 |
399.7 |
0.48 |
16.4 |
|
CA4 × DH6 |
6.6 |
0.37 |
1.7 |
10.2 |
0.36 |
2.1 |
237.3 |
0.32 |
10.1 |
|
CA4 × DH7 |
10.0 |
0.56 |
3.1 |
11.6 |
0.37 |
2.2 |
414.2 |
0.56 |
19.9 |
|
CA4 × DH8 |
9.0 |
0.46 |
2.4 |
10.9 |
0.38 |
2.2 |
374.2 |
0.48 |
15.5 |
|
CA5 × DH2 |
9.9 |
0.49 |
2.7 |
16.3 |
0.40 |
2.9 |
205.9 |
0.57 |
12.3 |
|
CA5 × DH3 |
6.6 |
0.31 |
1.4 |
9.2 |
0.40 |
2.1 |
227.4 |
0.56 |
8.1 |
|
CA5 × DH4 |
18.8 |
0.54 |
4.2 |
14.3 |
0.55 |
3.7 |
342.1 |
0.58 |
17.7 |
|
CA5 × DH5 |
9.4 |
0.59 |
3.2 |
6.7 |
0.47 |
2.1 |
566.0 |
0.52 |
24.8 |
|
CA5 × DH6 |
8.0 |
0.57 |
2.9 |
11.9 |
0.55 |
3.4 |
243.3 |
0.49 |
15.8 |
|
CA5 × DH7 |
9.9 |
0.62 |
3.4 |
9.9 |
0.32 |
1.7 |
289.6 |
0.54 |
18.6 |
|
CA5 × DH8 |
10.9 |
0.52 |
3.0 |
10.9 |
0.37 |
2.1 |
260.8 |
0.51 |
14.8 |
Among F4 populations, plant height varied between 143 to 183cm for CA2 × DH5 and CA4 × DH4, respectively with the mean value of 166cm (Table 2).
Significant variations were observed for plant height among all genotypes. On overall basis, variance for plant height ranged from 34.7 to 659.2 for CA5 and CA2 × DH8, respectively. Variances among parents ranges from 34.7 to 233.8 for genotypes CA5 and DH8, respectively while variances among F4 populations varied from 123.6 to 659.2 for CA2 × DH2 and CA2 × DH8, respectively. Moderate heritability in broad sense was observed for majority of the populations. The estimations of heritability ranged from 0.32 to 0.60 for CA4 × DH6 and CA2 × DH3, respectively. Genetic advance ranged from 8.6 to 25 for CA4 × DH6 and CA2 × DH8, respectively (Table 3). Most of the populations had moderate heritability coupled with high genetic advance, which shows that there is more genetic control over plant height therefore improvement in this character will be effective through selection. Our results are in line with the findings of Tariq et al. (2003) and Zhang and Zhou (2006) who reported high heritability and high genetic advance for plant height in Brassica napus and Brassica juncea, respectively.
Table 4: Variances, heritability and genetic advance of number of primary branches plant-1 and main raceme length in Brassica napus L.
|
Genotypes |
No. of Primary branches (nos) |
Main raceme length (cm) |
||||
|
Var. |
h2(bs) |
GA |
Var. |
h2(bs) |
GA |
|
|
CA2 |
0.8 |
24.0 |
||||
|
CA4 |
0.4 |
88.1 |
||||
|
CA5 |
0.8 |
27.4 |
||||
|
DH2 |
1.9 |
81.7 |
||||
|
DH3 |
0.6 |
83.4 |
||||
|
DH4 |
0.7 |
204.1 |
||||
|
DH5 |
0.6 |
147.7 |
||||
|
DH6 |
1.3 |
21.5 |
||||
|
DH7 |
0.9 |
43.6 |
||||
|
DH8 |
1.6 |
87.5 |
||||
|
CA2 × DH2 |
4.0 |
0.57 |
2.0 |
82.0 |
0.26 |
4.1 |
|
CA2 × DH3 |
1.4 |
0.43 |
0.9 |
331.6 |
0.44 |
14.1 |
|
CA2 × DH4 |
5.6 |
0.54 |
2.2 |
316.3 |
0.58 |
18.1 |
|
CA2 × DH5 |
1.6 |
0.55 |
1.2 |
174.6 |
0.43 |
10.1 |
|
CA2 × DH6 |
2.9 |
0.58 |
1.7 |
48.2 |
0.45 |
5.5 |
|
CA2 × DH7 |
3.9 |
0.57 |
2.0 |
57.7 |
0.40 |
5.4 |
|
CA2 × DH8 |
3.4 |
0.47 |
1.5 |
119.6 |
0.53 |
10.2 |
|
CA4 × DH2 |
3.2 |
0.61 |
1.9 |
141.7 |
0.40 |
8.4 |
|
CA4 × DH3 |
1.3 |
0.57 |
1.1 |
216.1 |
0.59 |
15.1 |
|
CA4 × DH4 |
1.4 |
0.60 |
1.3 |
406.0 |
0.53 |
18.8 |
|
CA4 × DH5 |
7.2 |
0.57 |
2.7 |
461.0 |
0.55 |
20.9 |
|
CA4 × DH6 |
2.7 |
0.58 |
1.7 |
119.7 |
0.50 |
9.6 |
|
CA4 × DH7 |
1.6 |
0.49 |
1.1 |
168.3 |
0.50 |
11.4 |
|
CA4 × DH8 |
2.5 |
0.52 |
1.5 |
253.9 |
0.48 |
13.3 |
|
CA5 × DH2 |
3.4 |
0.54 |
1.8 |
125.4 |
0.53 |
10.4 |
|
CA5 × DH3 |
2.3 |
0.58 |
1.5 |
75.9 |
0.34 |
5.2 |
|
CA5 × DH4 |
2.0 |
0.47 |
1.2 |
428.4 |
0.59 |
21.5 |
|
CA5 × DH5 |
1.7 |
0.61 |
1.4 |
274.3 |
0.54 |
15.6 |
|
CA5 × DH6 |
2.9 |
0.54 |
1.6 |
64.2 |
0.41 |
5.8 |
|
CA5 × DH7 |
1.8 |
0.53 |
1.3 |
90.2 |
0.59 |
9.8 |
|
CA5 × DH8 |
2.5 |
0.48 |
1.3 |
106.5 |
0.45 |
8.3 |
Primary branches plant-1
Number of primary branches is an important character of rapeseed and it is mainly related to the seed yield. Greater number of primary branches mostly produce greater number of pods that ultimately increase the seed yield in brassica. In current study, highly significant differences (P ≤ 0.01) among genotypes, parents, F4 populations and parents versus F4 for number of primary branches (Table 1) were observed. Similar findings have been reported earlier by Azadgoleh et al. (2009), Gangapur et al. (2009), Khan et al. (2013) and Ali et al. (2014). They also observed significant differences among brassica genotypes for number of primary branches.
Overall, mean value for primary branches plant-1 varied from 4 to 9 for CA2 × DH3 and CA2, respectively with the average value of 5.9 (Table 4). Number of primary branches among parents ranged from 5 (DH7) to 8 (CA2) with mean of 6 branches. Among F4 populations, primary branches plant-1 ranged from 4 (CA2 × DH3) to 8 (CA4 × DH4) with an average value of 6 branches plant-1 (Table 2).
Significant variations were observed among genotypes for primary branches indicating the presence of sufficient amount of variance among genotypes. Overall, variances among genotypes ranged from 0.4 to 7.2. Among parents, variances ranged from 0.4 to 1.9 for CA4 and DH2, respectively. Whereas among F4 populations, variances varied between 1.2 to 7.2 for CA4 × DH3 and CA4 × DH5, respectively. Heritability estimates for F4 populations exhibited moderate values for most of the populations. Heritability varied from 0.43 to 0.61 for cross combinations CA2 × DH3 and CA5 × DH5, respectively. Genetic advance ranged from 0.9 to 2.7, where minimum value was recorded for cross CA2 × DH3, while maximum value was noted for cross CA4 × DH5 (Table 4). Moderate heritability and genetic advance showed moderate genetic control suggesting that improvement in this trait possible through effective selection. Khan et al. (2013) and Ali et al. (2014) observed high heritability in Brassica napus genotypes. Similar results were earlier reported by Bozokalfa et al. (2010) in Eruca genotypes.
Main raceme length (cm)
Main raceme length is given due consideration in brassica breeding programs since longer racemes generally produce greater number of pods and ultimately enhancing seed yield. In the present study highly significant differences (P ≤ 0.01) were observed for main raceme length among the genotypes, parents and F4 populations whereas, significant differences (P ≤ 0.05) were observed among parents versus F4 (Table 1). Tahir et al. (2006) and Ali et al. (2014) also observed significant differences among brassica genotypes for main raceme length.
In the genotypes the minimum raceme length was noted for CA5×DH4 (47.1cm) while the maximum length was recorded for CA2 (80.9cm). Parental mean data showed that minimum main raceme length was recorded for DH8 (59.1cm) while maximum main raceme length was displayed by CA2 (80.9cm). Considering the F4 populations, the main raceme length ranged from 47.1cm to 77cm for CA5 × DH4 and CA2 × DH2, respectively. Average mean value of main raceme length was 65cm, 68cm and 64cm for genotypes, parents and F4 populations, respectively (Table 2).
Variance on overall basis ranged from 21.15 to 461cm for DH6 and CA4 × DH5, respectively. For parents, variance ranged from 21.5 to 204.07cm for DH6 and DH4, respectively. Among F4 populations, variances ranged from 48.23 to 460.97 for CA2 × DH6 and CA4 × DH5, respectively (Table 4). Moderate to high heritability were noted for majority of the crosses for main raceme length. The range of heritability among F4 population was 0.26 to 0.59 for CA2 × DH2 and CA5 × DH4, respectively. Genetic advance values ranged from 4.1 to 21.5 for CA2 × DH2 and CA5 × DH4, respectively (Table 4). Ali et al. (2014) also reported high heritability coupled with high genetic advance in Brassica napus. Zhang and Zhou (2006) reported similar results in Brassica oleracea.
Author’s Contribution
Tahir khan was the main investigator of the research work, while Raziuddin was the supervisor. Muazam Jamal Razi helped in data collection. Fakharuddin helped in data collection, analysis and preparation of 1st draft of the paper.
Acknowledgment
The authors are pleased to acknowledge HEC, Pakistan for sponsoring this research under project No. 20-1377 under NURSP.
References
Azadgoleh, E.M.A., M. Zamani and E. Yasari. 2009. Agronomical important traits correlation in rapeseed genotypes. Res. J. Agri. Biol. Sci. 5(5):798-802.
Ahmad, R., Farhatullah and C.F. Quiros. 2011. Inter and intra-cluster heterosis in spring type oilseed rape (Brassica napus L.) hybrids and prediction of heterosis using SRAP molecular markers. Sabrao J. Breed. Gen. 43(1):27-43.
Ahmad, M., M. Naeem, I.A. Khan, Farhatullah and M.N. Mashwani. 2012. Biochemical quality study of genetically diversified brassica genotypes. Sarhad J. Agri. 28(4):599-602.
Azam, S.M., Farhatullah, A. Nasim, S. Shah and S. Iqbal. 2013. Correlation studies for some agronomic and quality traits in Brassica napus L. Sarhad J. Agri. 29(4):547-550.
Bozokalfa, K.M., D.E. Ilbi and T.K. Asciogul. 2010. Estimates of genetic variability and association studies in quantitative plant traits of Eruca spp. Landraces. Genetika. 42(3):501-512. http://dx.doi.org/10.2298/GENSR1003501B
Chaghakaboodi, Z., D. Kahrizi and A. Zebarjadi. 2012. Heritability and genetic advance in rapeseed (Brassica napus L.). Iran. J. Gen. Plant Breed. 1(2):16-21.
Falconer, D.S., and T.F.C. Mackay. 1996: Introduction to Quantitative Genetics (4th Ed.) Longman, Essex, UK.
FBS. 2014. Federal Bureau of Statistics. The government of Pakistan.
FAO. 2014. Food and Agricultural Organization United Nation.
Gangapur, D.R., B.G. Prakash, P.M. Salimath, R.L. Ravikumar and M.S.L. Rao. 2009. Correlation and path analysis in Indian mustard. Karnataka J. Agri. Sci. 22(5):971-977.
Hasan, E., H.S.B. Mustafa, T. Bibi, and M. Mahmood. 2014. Genetic variability, correlation and path analysis in advanced lines of rapeseed (Brassica napus l.) for yield components. Cercetari Agronomicein Moldova. 47(1):71-79.
Jatoi, S.A., A. Javaid, M. Iqbal, O.U. Sayal, M. S. Masood and S.U. Siddiqui. 2012. Genetic diversity in radish germplasm for morphological traits and seed storage proteins. Pak. J. Bot. 43(5):2507-2512.
Khan, F.A., S. Ali, A. Saeed and G. Abbas. 2006. Genetic variability and genetic advance analysis for some morphological traits in B. napus L. J. Agri. Res. 44(2):83-88.
Khan, F.U., Raziuddin, I.A. Khalil, I.H. Khalil and Iltafullah. 2013. Heritability and genetic potential of Brassica napus genotypes for yield and yield components. American-Eurasian J. Agri. Environ. Sci. 13(6):802-806.
Mahmud, I., and Kramer, H.H. 1951. Segregation for yield, height and maturity following a soybean cross. Agron. J. 43:605-609. http://dx.doi.org/10.2134/agronj1951.00021962004300120005x
Mahto, J.L., and Z.A. Haider. 2002. Variability studies in Indian mustard under six different environments in acidic soil. Crucif Newsletter. 24:13-14.
Marinkovic, R., A.M Jeromela and D. Vasic. 2003. Genetic variability components of some quantitative traits of winter oilseed rape (Brassica napus L.). Genetika. 35(3):199-205. http://dx.doi.org/10.2298/GENSR0303199M
Marwede, V., A. Schierholt and H.C. Becker. 2004. Genotype x environment interactions and heritability of tocopherol contents in canola. Crop Sci. 44:728-731. http://dx.doi.org/10.2135/cropsci2004.0728
Mahmood, T., M. Hussain, M.S. Tahir, M. Sharif and Tahira. 2011. Punjab sarson: An introduction of new canola version high yielding variety released for general cultivation in the Punjab. Pak. J. Agri. Sci. 48:263-267.
Muhammad, A., Raziuddin, A. Muhammad, H. Raza, A. Rahman and I. Ali. 2014a. Combining ability and heritability studies for important traits in F2 populations of Brassica napus L. Int. J. Basic Appl. Sci. 14(01):7-11.
Ali, M., Raziuddin, M. Sajid, A. Rahman and S.A. Khan. 2014. Combining ability and heritability studies for yield contributing traits in F2 populations of Brassica napus L. Am. J. Agri. Environ. Sci. 14(06):509-515.
Nazeer, A., F. Javidfar, J.Y. Elmira and M. Mirza. 2003. Relationship among yield components and selection criteria for yield improvement in winter rapeseed (Brassica napus L.). Pak. J. Bot. 35:167-174.
Nasim, A., Farhatullah, S. Iqbal, S. Shah and S. M. Azam. 2013. Genetic variability and correlation studies for morpho-physiological traits in Brassica napus L. Pak. J. Bot. 45(4):1229-1234.
Panse, V.G., and P.V. Sukhatme. 1965: Statistical Methods for Agricultural Workers. Indian Council of Agriculture Research. New Dehli. p. 381.
Rameeh, V. 2014. Correlation and factor analyses of quantitative traits in rapeseed (Brassica napus L.). Int. J. Agri. Innov. Res. 1(1):2319-1473.
Steel, R.G.D., and J.H. Torrie. 1980: Principles and procedures of statistics, a biological approach, 2nd ed. McGraw Hill, Inc. New York, Toronto, London.
Syed, A.S., I. Ali and K. Rahman. 1994. Mutation breeding of oil seed crops, proceedings of a final research co-ordination meeting of an FAO/IAEA Coordinated Research Program, Vienna, 11-15 Jan. 1993. 81:25-35.
Singh, M., G.B. Swarnker, I. Prasad and G. Rai. 2002. Genetic variability, heritability and genetic advance for quality traits in Indian mustard. Plant Arch. 2(1):27-31.
Sabaghnia, N., H. Dehghani, B. Alizadeh and M. Mohghaddam. 2010. Heterosis and combining ability analysis for oil yield and its components in rapeseed. Aust. J. Crop Sci. 4(6):390-397.
Tariq, M., A. Muhammad, I. Shahid and A. Muhammad. 2003. Genetic variability and heritability estimates in summer mustard. Asian J. Plant Sci. 2(1):77-79. http://dx.doi.org/10.3923/ajps.2003.77.79
Tahir, M.H.N., S. Bashir, and A. Bibi. 2006. Genetic potential of canola varieties under water stress conditions. Caderno de Pesquisasér. Bio.logic. 18(2):127-135.
Zhang, G., and W. Zhou. 2006. Genetic analyses of agronomic and seed quality traits of synthetic oilseed B. napus produced from inter specific hybridization of Brassica campestris and Brassica olearacea. J. Gen. 85(1): 45-51. http://dx.doi.org/10.1007/BF02728969
Zare, M., and S. Sharafzadeh. 2012. Genetic variability of some rapeseed (Brassica napus L.) cultivars in Southern Iran. African J. Agri. Res. 7(2):224-229.
To share on other social networks, click on any share button. What are these?








