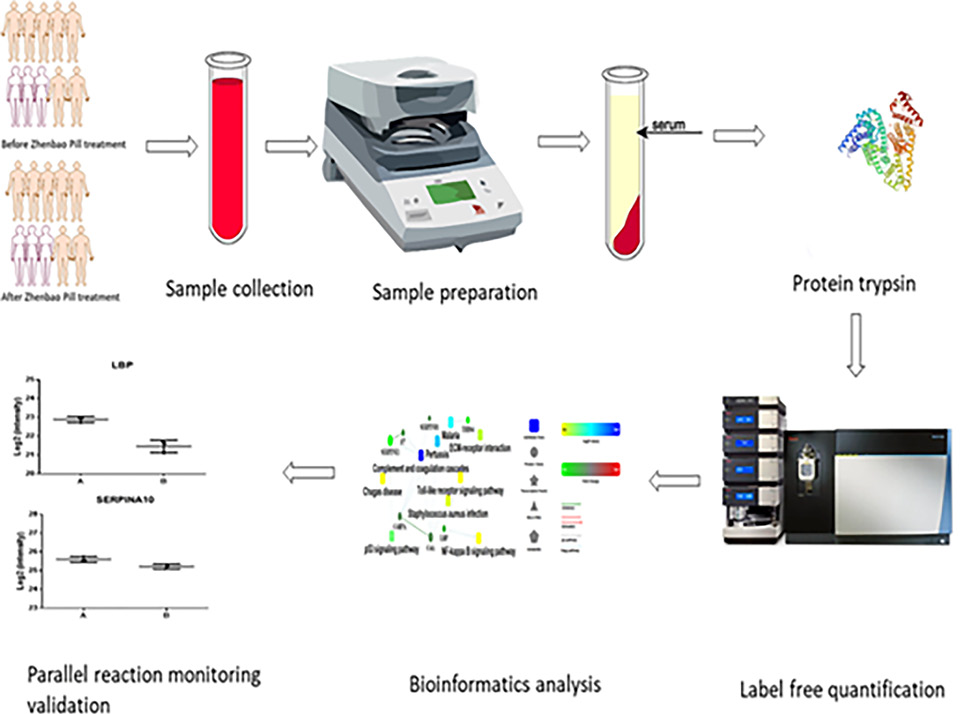
Label-Free Quantification based Proteomic Analysis of Serum Obtained from Henoch–Schönlein Purpura Patients before and after Zhenbao Pill Treatment
Label-Free Quantification based Proteomic Analysis of Serum Obtained from Henoch–Schönlein Purpura Patients before and after Zhenbao Pill Treatment
Wuritunashun*, Burenbatu, Narenqiqige, Jiuguniang, Eerdunduleng, Shuanglian Wang, Cuiqin Gong, Hashengaowa, Huizhi Jin, Baiwurihan and Chunhaizi
(A) Summary of the total number of GO hits. (B) Total number of upregulated and downregulated proteins using LFQ analysis.
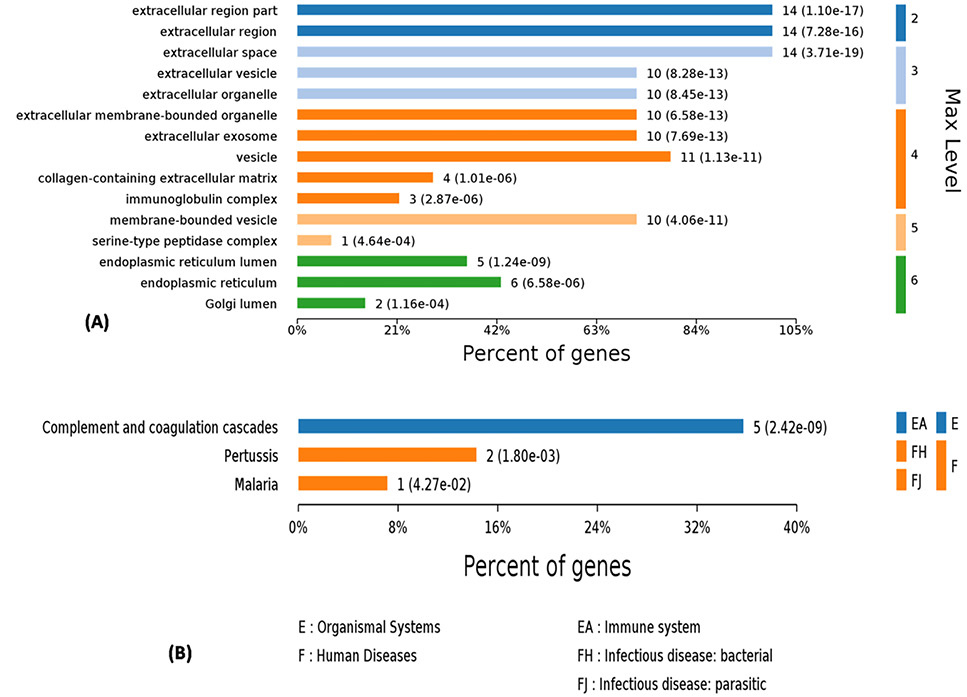
Biological processes involving the differentially expressed proteins. Each bar includes the number of genes and p-value calculated with Fish exact test with Hypergeometric algorithm. Max level means maximal annotated level of this term in the GO graph.
(A) Cellular components, (B) KEGG pathways involving the differentially expressed proteins.
Protein protein interactions network of the differentially expressed proteins.
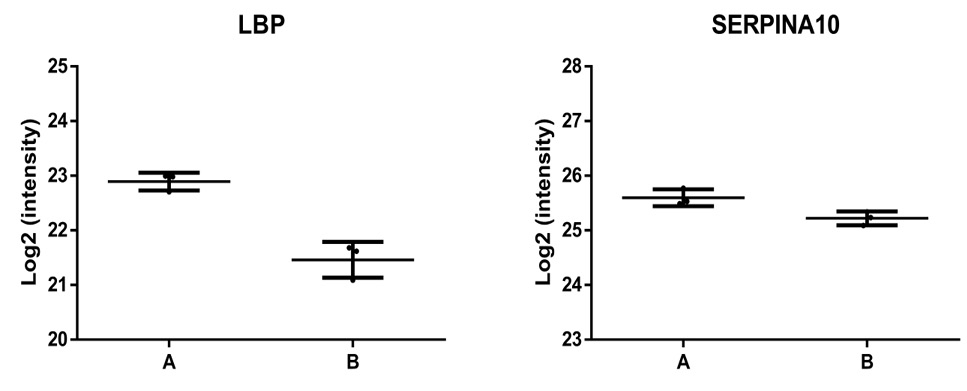
Validation of differentially expressed proteins (DEPs) using parallel reaction monitoring analysis. Two candidate DEPs were used to verify the LFQ proteomics results.