Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (2): 91 – 97VP5 Gene Based Molecular Comparison of Indian and Global Isolates of Bluetongue Virus 2
Koushlesh Ranjan, Prasad Minakshi*, Pawan Kumar, Gaya Prasad
*Corresponding author: [email protected]
ARTICLE CITATION:
Ranjan K, Minakshi P, Kumar P, Prasad G. (2014). VP5 gene based molecular comparison of Indian and global isolates of bluetongue virus 2. Adv. Anim. Vet. Sci. 2 (2): 91 – 97.
Received: 2013–12–12, Revised: 2013–12–20, Accepted: 2013–12–21
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.2.91.97
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Bluetongue virus (BTV) is a prototype species of the genus Orbivirus within family Reoviridae which causes bluetongue (BT) disease in domestic as well as wild ruminants. BTV is non–enveloped virus having 10 segmented dsRNA genome. Segment 6 encodes VP5 protein which along with VP2 protein gives serotype specificity to the virus. The BT2 isolate (Accession number JF899598) was grown in BHK 21 cell culture. The viral dsRNA was extracted from cell culture material. A pair of vp5 gene specific primer (Forward primer 288–307 bp and reverse primer 738–757 bp) giving an amplicon size of 470bp was designed using vp5 gene sequences available in GenBank database. The primer pair specifically amplified the desired region of template. The BT2 isolate showed maximum nucleotides (99.5–99.7%) as well as deduced amino acids (98.7–99.3%) sequence identity with South African, USA and most of the Indian isolates of BTV2 serotype. The sequence identity with European, Sudanese and Nigerian isolates was 85.7–87.2% for nucleotides and 94.2–95.5% for deduced amino acids. However, it showed 76.8–77.8% nucleotide and 87.1–87.8% deduced amino acid identity with eastern isolates. Results indicated the western origin of BT2 isolate (Accession number JF899598). Moreover, phylogenetic analysis also revealed the close relationship of BT2 isolate with Western BTV2 isolates. In–silico restriction enzyme analysis revealed that isolate in study can be differentiated from other BTV2 isolates from India, USA, South Africa, Europe, Australia and Japan using HindII, AarI, AcyI, AflIII, BsmAI restriction enzyme.
INTRODUCTION
Bluetongue (BT) is an economically important non–contagious, infectious, insect vector (Culicoides) borne disease of domestic and wild ruminants (Maclachlan et al., 1994). It is caused by BT virus (BTV) of genus Orbivirus and family Reoviridae. BT causes considerable economic loss to livestock industry which is mainly attributed to high morbidity, mortality, abortion, still birth, foetal abnormality, weight loss, wool break, reduced milk and meat yield. BT is listed as a multiple species diseases by Office International des Epizooties (OIE, 2013). It causes severe clinical signs as evidenced by fever, lameness, swelling of lips and tongue. The more severe forms of the disease are frequently seen in sheep and in white–tailed deer (Howerth et al., 1988; Darpel et al., 2007). Buffalo, cattle and goats act as silent reservoirs and remain viraemic for several months (Maclachlan et al., 2009).
Due to rapid evolutionary changes in genome, through reassortment and mutations, BTVs are consistently evolving new serotypes globally. Twenty–four distinct serotypes (BTV1 to BTV24) of the virus have been identified worldwide (Mertens et al., 2004). However, recently BTV25 from Switzerland (Hofmann et al., 2008) and BTV26 from Kuwait have been isolated (Maan et al., 2011). Since India is endemic for Culicoides vector, a total of 21 BTV serotypes have been reported so far (Prasad et al., 2007). Recently, the 22nd BTV serotype i.e. BTV21 have been reported from Andhra Pradesh state (Susmitha et al., 2012). Based on nucleotide sequence analysis, most of BTV isolates can be broadly divided into two major groups ‘eastern’ or ‘western’ topotypes, and into a number of geographic subgroups (Balasuriya et al., 2008). BTV is an icosahedral virus with a ten–segmented, double stranded RNA (dsRNA) genome. Each of the ten segments codes for at least one viral protein. Seven proteins (VP1 to VP7) are structural and form virus particle. In addition to structural proteins, there are four non–structural proteins NS1, NS2, NS3 and NS3a expressed in virus infected host cells (Mertens et al., 1989). Recently, it has been identified that segment 9 encodes another non–structural protein (NS4) (Ratinier et al., 2011; Belhouchet et al., 2011). The inner capsid of BTV is composed of five polypeptides: three minor proteins (VP1, VP4, and VP6) and two major proteins (VP3 and VP7) (Roy, 1989). The outer capsid is composed of serotype specific two viral proteins, VP2 and VP5 (Ghiasi, 1987).
VP5 protein participates in virus neutralization activity along with VP2 protein as it enhances the neutralization activity of VP2 protein (Roy et al., 1990). Combination of soluble VP2 and VP5 proteins can induce higher neutralizing antibody titre than VP2 protein alone (Huismans et al., 1983). In addition, VP5 also plays an important role in membrane permeabilization and membrane fusion activity facilitating virus entry into the host cells (Forzan et al., 2004). The BTV2 has been reported earlier from India (Balumahendrian et al., 2009; Maan et al., 2012a,b). The present study was carried out for vp5 gene based molecular characterization and for determining the phylogenetic relationship of Indian isolate of BTV2 with global isolates.
MATERIALS AND METHODS
Sample Origin
The virus isolate was obtained under All India Network Programme on Bluetongue. The virus isolate was originally isolated from sheep blood (Nellore breed) in 2004 from Andhra Pradesh state and designated as isolate BT2. The virus sample was propagated in BHK–21 cell line in our lab.
Isolation of Viral dsRNA
The BT2 isolate produced BTV specific cytopathic effect (CPE) in infected BHK–21 cell culture within 48 hours. After appearance of about 75% CPE in infected BHK–21 cell culture, the virus was harvested. The viral dsRNA was extracted using TRIzol reagent (Invitrogen, USA) as per manufacturer’s instructions.
Oligonucleotide Primers
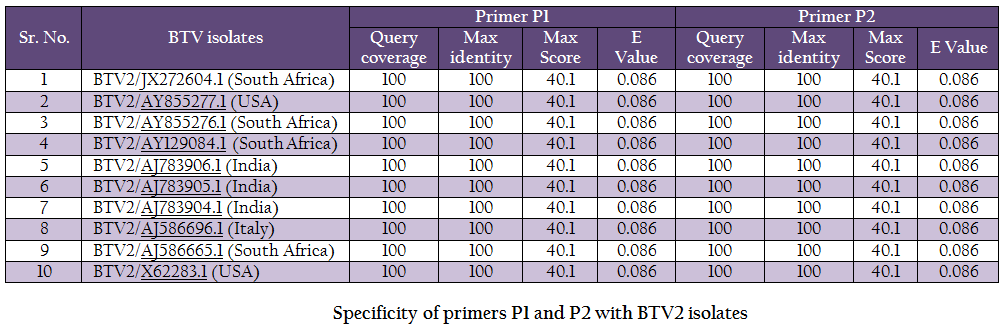
The vp5 gene specific primers P1 (288–307nt) – 5’TGAGCCCGGGTGAGCAAGGT 3’ and P2 (738–757nt) – 5’TGTCGCCATACCCGCTCCGA3’ with expected amplicon size of 470bp were designed based on nucleotide sequences of serotype BTV2 available in GenBank with the help of Primer Blast software (http://www.ncbi.nlm.nih.gov/tools/primer–blast/). Upon BLASTn similarity search the primer pairs P1 and P2 showed significant similarity with BTV2 isolates in GenBank data base only (Table 1). It showed its specificity to BTV2 serotypes. The primer sequences were got synthesized commercially (LifeTech, USA).
Polymerase Chain Reaction (PCR)
The viral genomic dsRNA was used as a template for cDNA synthesis using random primers (decamer) (Ambion, USA) and moloney murine leukemia virus–reverse transcriptase
(Mo–MuLV–RT) enzyme (Sibzyme, Russia). The reaction mixture consisted of 7µg of heat denatured viral dsRNA, 6% DMSO and 30 pMol of random decamer. Finally, 25 µl reaction mix was prepared using 400µM each dNTPs and 500 U of Mo–MuLV reverse transcriptase (Promega, USA) for reverse transcription. The primer was allowed to anneal at 250C for 10 min followed by reverse transcription at 370C for 60 min and inactivation for 10 min at 900Cin thermal cycler (Biorad i–Cycler, USA). The cDNA was subjected to PCR with group specific NS1 gene based primer to confirm the sample as BTV using standard protocol (Kovi et al., 2005). The cDNA was subsequently amplified using primer pair P1 and P2 along with vp5 gene specific primers of all serotypes. The amplification was carried out in 20 µl reaction mixture containing 2 µl cDNA, 3% DMSO, 20 µM of primers P1 and P2 along with 0.4 µl of 10mM dNTPs mix, 4 µl 5X HF buffer and 0.4 U (2U/µl) phusion high– fidelity DNA polymerase (Finnzymes, Finland) in a thermal cycler (Biorad iCycler, USA). The amplification programme consisted of initial denaturation for 3 min at 980C, followed by 35 cycles for 10 sec denaturation at 980C, 20 sec primer annealing at 570C and 30sec primer extension at 720C. Final primer extension was carried out at 720C for 10 min. The amplified products were analysed by electrophoresis using 1% agrose gel (Sigma, USA) stained with ethidium bromide and visualized under UV trans illuminator (Biovis, USA).
VP5 Gene Cloning and Sequencing
PCR products were purified using QIA quick gel extraction kit (Qiagen, USA) to remove primer dimmers and other PCR ingredients. The purified PCR products were cloned using commercial kit (Fermentas, USA) in Pjet 1.2 cloning vector as per the manufacturer’s instruction. The JM107 cell line was used as host system. The positive clones were selected by colony touch PCR using vp5 gene specific primer pair. The plasmids from positive clones were extracted using commercial kit Quiaprep (Quiagen, USA) as per the manufacturer’s instruction.
Nucleotide Sequencing
The plasmids from positive clones were allowed for nucleic acid sequencing using vector specific primer by Genetic Analyser ABI PRISM TM 3130 XL machine in our laboratory.
Nucleotide Sequence Analysis
The vector sequence from nucleotide sequence generated was removed using Vecscreen software (http: //www. ncbi. nlm.nih. gov /tools /vecscreen/). The vp5 gene sequence of BT2 isolate was analysed using online software BLASTNn 2.2.28 (http://blast.ncbi.nlm.nih.gov/) (Zhang et al., 2000). Bioedit v7.2.3 software was used for multiple sequence alignment and calculation of percent identity of vp5 gene of Indian BTV2 isolate with other global isolates. Similarly, the percent identity of deduced amino acid sequence of vp5 gene was calculated with global isolates using Bioedit v7.2.3 software. The nucleotide as well as deduced amino acid based phylogenetic analysis of BT2 isolate with other isolates of BTV2 from different parts of the world was done using neighbour joining algorithm of Mega 5 software (Tamura et al., 2011). The neighbour joining tree was constructed using p–distance parameter with bootstrap value 1000. To study the genomic diversity, the in–silico restriction enzyme analysis of vp5 gene of isolate in study with BTV2 global isolates were carried out using Restriction Mapper version 3.0 software (http://www .restrictionmapper.org/).
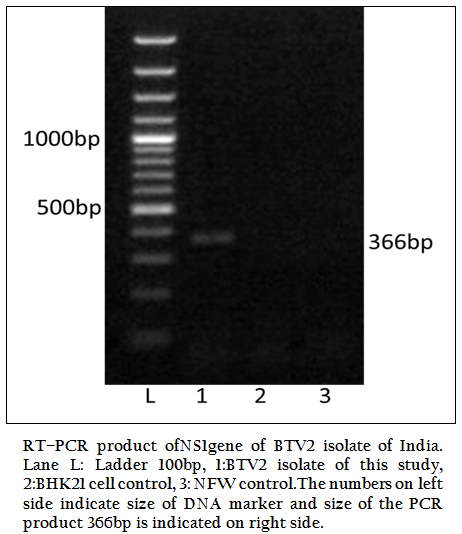
Figure 1: RT–PCR product ofNS1gene of BTV2 isolate of India. Lane L: Ladder 100bp, 1:BTV2 isolate of this study, 2:BHK21 cell control, 3: NFW control.The numbers on left side indicate size of DNA marker and size of the PCR product 366bp is indicated on right side.
RRESULTS
The BT2 isolate was propagated in BHK21 cell line. It showed BTV specific cytopathic effect (CPE) such as cellular aggregation, foamy degeneration and detachment of cells from surface within 48 hours. The viral nucleic acid (dsRNA) was extracted and cDNA was prepared. The group specific NS1 gene based PCR showed 366bp amplicon size in 1% agarose gel electrophoresis (Figure 1). The BTV specific CPE in BHK–21 cells and specific amplification of NS1 gene (366bp) confirmed the sample as BTV. The vp5 gene of BT2 isolate was specifically amplified by BTV2 specific primer pairs (P1 and P2) as evidenced by expected 470bp amplicon size of PCR product (Figure 2). The remaining serotype specific primers did not show any amplification (data not shown). The primer pairs P1 and P2 specific amplicon was cloned and sequenced.
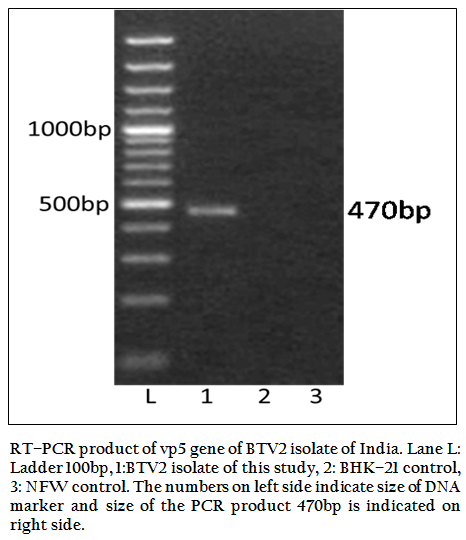
Figure 2: RT–PCR product of vp5 gene of BTV2 isolate of India. Lane L: Ladder 100bp, 1:BTV2 isolate of this study, 2: BHK–21 control, 3: NFW control. The numbers on left side indicate size of DNA marker and size of the PCR product 470bp is indicated on right side.
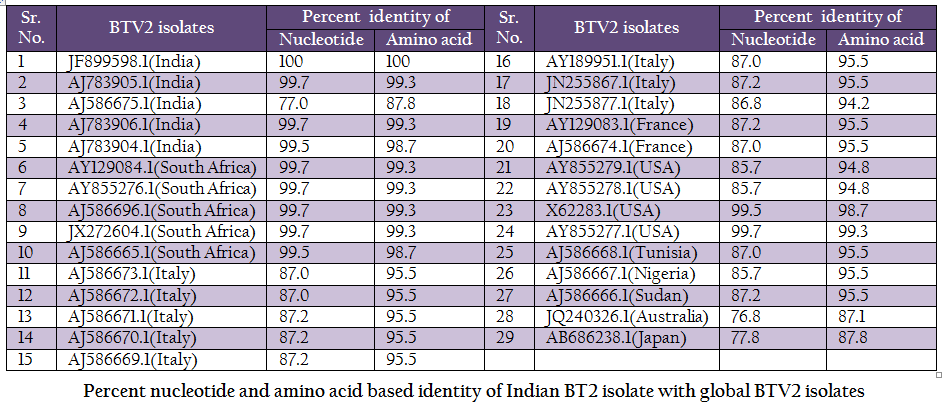
Table 2: Percent nucleotide and amino acid based identity of Indian BT2 isolate with global BTV2 isolates.
DISCUSSION
BTV is endemic in India and, in recent past eight different BTV serotypes have been isolated there (Rao et al., 2012). BTV2 is one of the common serotype reported from India (Balumahendrian et al., 2009). The significant genetic diversity (3–43%) has been observed based on vp5 gene sequences of all the 24 serotypes of BTV across the world (Singh et al., 2004). The vp5 gene based molecular characterization and phylogenetic relationship of Indian BTV serotype 1 (Manjunath et al., 2010) and reference strains of all 24 serotypes (Singh et al., 2004) have been reported earlier. In present study, the new primer pair was designed for the amplification of vp5 gene of Indian BTV2 isolate. The newly designed primer specifically amplified BTV2 isolate. The primer pair was tested with blastn search of GenBank data base and it showed significant similarity with BTV2 isolates only (Table 1).
The BT2 isolate (Accession number JF899598) was closely related with Indian, South African and USA isolates of BTV2. It showed maximum identity of nucleotide (99.7%) and amino acid (99.3%) with reference western BTV2 isolate from Tamil Nadu, India (Accession number AJ783905) (Maan et al., 2012a). It also showed higher nucleotide as well as amino acid identity with other western BTV2 isolates from India, South Africa and USA. The isolate in study was distantly related (85.7–87.2% nucleotide and 94.2–95.5% amino acid identity) to European, Sudan and Nigeria isolate. However, it was far distantly related (nucleotide 77.0% and amino acid 87.8%) with one of the Indian BTV2 isolate (Accession number AJ586675.1) (Maan et al., 2012b). Moreover, it was distantly related (76.8–77.8% nucleotide and 87.1–87.8% amino acid identity) with other eastern BTV2 isolate from Australia and Japan. This indicated that the BT2 isolate was closer to western topotype of BTV2. Further, sequence analysis showed only little diversity in nucleotide (0–0.4%) as well as deduced amino acid (0–0.6%) among closely related Indian BTV2 isolates of western origin (Accession no. AJ783904, AJ783905 and AJ783906) and isolate in study (Accession number JF899598). It indicated the common origin of Indian western BTV2 isolates.
The phylogenetic studies of nucleotide as well as deduced amino acid sequences of BT2 isolate (Accession number JF899598) along with other global BTV2 isolates were calculated. The result showed that the BTV2 isolates were grouped in two major clusters of western and eastern origin. The major western cluster was sub grouped in three sub clusters I, II, and III (Figure 3 and Figure 4). Based on nucleotides and deduced amino acids phylogenetic analysis, the BT2 isolate (Accession number JF899598) and most of the USA, South Africa and Indian isolates were placed in sub cluster II of western cluster. The BTV2 from Europe, Nigeria and Sudan were grouped in separate sub cluster I of western cluster. The two USA isolates (Accession number AY855278 and AY855279) were placed in separate sub cluster III of western cluster. The major eastern cluster was formed with one of the Indian (Accession no. AJ586675), Australian and Japanese isolates. Thus the phylogenetic analysis also indicated the western origin of BT2 isolate.
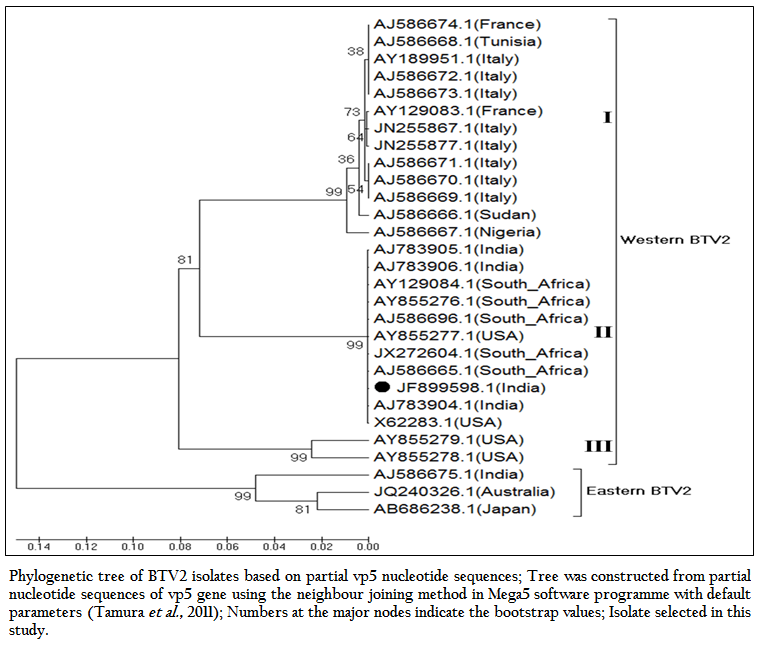
Figure 3: Phylogenetic tree of BTV2 isolates based on partial vp5 nucleotide sequences; Tree was constructed from partial nucleotide sequences of vp5 gene using the neighbour joining method in Mega5 software programme with default parameters (Tamura et al., 2011); Numbers at the major nodes indicate the bootstrap values; Isolate selected in this study.
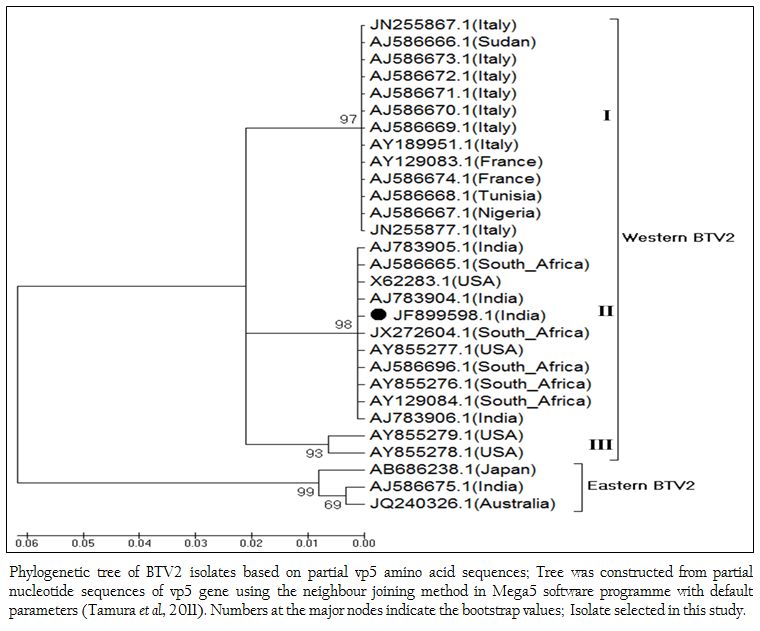
Figure 4: Phylogenetic tree of BTV2 isolates based on partial vp5 amino acid sequences; Tree was constructed from partial nucleotide sequences of vp5 gene using the neighbour joining method in Mega5 software programme with default parameters (Tamura et al., 2011). Numbers at the major nodes indicate the bootstrap values; Isolate selected in this study.
RT–PCR in combination with restriction analysis has been found very useful tool in molecular characterization of BTV isolates directly from clinical samples. In the present study, based upon HindII, AarI, AflIII, AcyI and BsmAI restriction enzyme sites, the BT2 (Accession number JF899598) isolate could be differentiated from European, Nigerian, Sudanese and eastern topotype isolates. The in–silico restriction enzyme analysis (REA) with HindII, AarI, AflIII and BsmAI revealed a single restriction site at 536, 400, 577 and 645 respectively in sub cluster II of western cluster isolates (isolate in study along with in most of the Indian, South Africa and USA isolates). However, sub cluster I isolates of western cluster (Sudan, Nigeria and European isolates) did not show and restriction site for enzymes HindII, AarI and AflIII. They showed two restriction sites at 578 and 645 with restriction enzyme BsmAI. The sub cluster III (USA isolates, Accession nos. AY855278 and AY855279) did not show any restriction sites for enzymes HindII, AarI, AflIII and BsmAI. They showed only one restriction site at 716 with restriction enzyme AcyI. Remaining all other BTV2 isolates did not show any restriction site with enzyme AcyI. The BTV2 isolates of eastern cluster (one of the Indian isolate (Accession no. AJ586675), Australian and Japan) revealed a single restriction site at 578 with BsmAI restriction enzyme. They did not show any restriction site with enzymes HindII, AarI, AflIII and AcyI. Based upon HindII, AarI, AflIII, AcyI and BsmAI restriction sites, appreciable genomic diversity was observed between BT2 isolate (Accession number JF899598) and the global BTV2 isolates. These findings provide the basis for differentiation of Indian isolate from European, USA, Nigerian Sudanese, Australian and Japan BTV2 isolates.
The prevalence of BTV2 from Tamil Nadu state (adjoining state of Andhra Pradesh) was documented earlier (Maan et al., 2012a). A higher percentage of nucleotide (99.7%) as well as deduced amino acid (99.3%) sequence identity was observed between the BT2 isolate (Accession number JF899598) and Tamil Nadu isolate (Accession no. AJ783905.1). Since Andhra Pradesh and Tamil Nadu are cross border States and are endemic for a major species of vector i.e. Culicoides oxystoma, the BTV2 serotype might have migrated from Tamil Nadu to Andhra Pradesh or vice versa either through vector or migrating sheep population, however, future studies are required to confirm such prediction.CONCLUSIONS
Based on percent nucleotide as well as deduced amino acid sequence identity of vp5 gene, it can be concluded that Indian BT2 isolate (Accession number JF899598) is more closely related to western topotype of BTV2 from India, USA and South Africa. The analyses of vp5 gene sequence revealed that there is no significant divergence found among Indian western BTV2 isolates. However based upon in–silico REA, Indian BTV2 isolates can be differentiated from European, Nigerian, Sudan and eastern topotype BTV2 isolates. RT–PCR along with in silico restriction analysis of partial vp5 gene of the Indian isolate could be helpful in serotyping of BTV. The newly designed primer pair for vp5 gene can be used for vp5 gene identification and molecular characterization of BTV serotype 2.
ACKNOWLEDGEMENTS
The study reported in present paper was funded by Indian Council of Agricultural Research, New Delhi under ‘All India network programme on Bluetongue’. The authors are thankful to the Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar for providing infrastructural facility.
CONFLICT OF INTEREST
We declare no conflict of interest.
REFERENCES
Balasuriya UBR, Nadler SA, Wilson WC, Pritchard LI, Smythe AB, Savini G, Monaco F, De Santis P, Zhang N, Tabachnick WJ, MacLachlan NJ (2008). The NS3 proteins of global strains of bluetongue virus evolve into regional topotypes through negative (purifying) selection. Vet. Microbiol. 126: 91 – 100.
http://dx.doi.org/10.1016/j.vetmic.2007.07.006
PMid:17706379
Balumahendrian M, Sreenivasulu D, Kumar CA, Suryanarayana VVS, Byregowda SM (2009). Characterization of vp2 gene of Indian bluetongue virus serotype 2 and its close phylogenetic relationship to the Taiwan isolate. Vet. Res. 86: 332 – 338.
http://dx.doi.org/10.1016/j.rvsc.2008.05.017
PMid:18649903
Darpel KE, Batten CA, Veronesi E, Shaw AE, Anthony S, Bachanek–Bankowska K, Kgosana L, Bin–Tarif A, Carpenter S, Müller–Doblies UU, Takamatsu HH, Mellor PS, Mertens PPC, Oura CAL (2007). A study of British sheep and cattle infected with bluetongue virus serotype 8 from the 2006 outbreak in northern Europe. Vet. Rec. 161: 253 – 261.
http://dx.doi.org/10.1136/vr.161.8.253
PMid:17720961
Forzan M, Wirblich C, Roy P (2004). A capsid protein of non–enveloped bluetongue virus exhibits membrane fusion activity. Proc. Natl. Acad. Sci. 101(7): 2100 – 2105.
http://dx.doi.org/10.1073/pnas.0306448101
PMid:14762165 PMCid:PMC357058
Ghiasi H, Fukusho A, Eshita Y, Roy P (1987). Identification and characterization of conserved and variable regions in the neutralization VP2 gene of bluetongue virus. Virol. 160: 100 – 109.
http://dx.doi.org/10.1016/0042-6822(87)90050-X
Howerth EW, Greene CE, Prestwood AK (1988). Experimentally induced bluetongue virus infection in white tailed deer: coagulation, clinical pathologic and gross pathologic changes. Am. J. Vet. Res. 49: 1906 – 1913.
PMid:2854709
Hofmann MA, Renzullo S, Mader M, Chaignat V, Worwa G, Thuer B (2008). Genetic characterization of Toggenburg orbivirus a new bluetongue virus from goats Switzerland. Emerg. Infect. Dis. 121: 1855 – 1861.
http://dx.doi.org/10.3201/eid1412.080818
PMid:19046507 PMCid:PMC2634640
Huismans HNT, Vander Walt, Cloete M, Erasmus BJ (1983). The biochemical and immunological characterization of bluetongue virus outer capsid polypeptides, In R.W. Compans and D. Bishop (ed.), Double–stranded RNA viruses (Elsevier, New York, N.Y), 165 – 172.
Susmitha B, Sudheer D, Rao PP, Uma M, Prasad G, Minakshi P, Hegde NR, Reddy YN (2012). Evidence of bluetongue virus serotype 21 (BTV–21) divergence. Virus Genes. 44(3): 466 – 469.
http://dx.doi.org/10.1007/s11262-012-0724-y
PMid:22350945
Kovi RC, Dahiya S, Minakshi, Prasad G (2005). Evaluation of new primers targeting serogroup specific genes for detection of bluetongue viruses by RT–PCR. Indian J. Microbiol. 45(2): 103 – 119.
Maan NS, Maan S, Guimera M, Pullinger G, Singh KP, Nomikou K, Belaganahalli MN, Mertens PPC (2012a). Complete Genome Sequence of an Isolate of Bluetongue Virus Serotype 2, Demonstrating Circulation of a Western Topotype in Southern India. J. Virol. 86(9): 5404 – 5405. doi:10.1128/JVI.00420–12.
http://dx.doi.org/10.1128/JVI.00420-12
Maan NS, Maan S, Nomikou K, Guimera M, Pullinger G, Singh KP, Belaganahalli MN, Mertens PPC (2012b). The Genome sequence of bluetongue virus type 2 from India: evidence for reassortment between eastern and western topotype field strains. J. Virol. 86(10): 5967 – 5968. DOI: 10.1128/JVI.00536–12.
http://dx.doi.org/10.1128/JVI.00536-12
Maan S, Maan NS, van Rijn PA, van Gennip RGP, Sanders A, Wright IM, Batten C, Hoffmann B, Eschbaumer M, Oura CAL, Potgieter AC, Nomikou K, Mertens PPC (2010). Full genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains. PLoS ONE. 5(4): 1 – 17.
http://dx.doi.org/10.1371/journal.pone.0010323
PMid:20428242 PMCid:PMC2859060
MacLachlan NJ (1994). The pathogenesis and immunology of bluetongue virus infection of ruminants. Comp. Immunol. Microbiol. Infect. 17: 197 – 206.
http://dx.doi.org/10.1016/0147-9571(94)90043-4
Maclachlan NJ, Drew CP, Darpel KE, Worwa G (2009). The pathology and pathogenesis of bluetongue. J. Comp. Path. 141: 1 – 16.
http://dx.doi.org/10.1016/B978-012369368-6.50017-4
Manjunath BN, Prasad M, Maan S, Prasad G (2010). Differentiation of Indian isolates of bluetongue virus serotype 1 from Australian and African isolates based on analysis of vp5 gene. Ind. J. Biotech. 9: 117 – 125.
Maan S, Maan NS, Nomikou K, Veronesi E, Bachanek–Bankowska K, Manjunatha BN, Attoui H, Mertens PPC (2011). Complete Genome Characterisation of a Novel 26th Bluetongue Virus Serotype from Kuwait. PLoS ONE 6(10): e26147. doi:10.1371/journal.pone.0026147
http://dx.doi.org/10.1371/journal.pone.0026147
OIE (2013). OIE–Listed diseases, infections and infestations in force in 2013. http://www.oie.int/animal-health-in-the-world/oie-listed-diseases-2013/.
Mertens PPC, Maan S, Samuel A, Attoui H (2004). Orbivirus, Reoviridae. In Virus Taxonomy, VIIIth Report of the ICTV, pp. 466– 483. Edited by C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger and L. A. Ball. London: Elsevier/Academic Press.
Mertens PPC, Pedly S, Cowly J, Burrough JN, Corteyn AH, Jeggo MH, Jennings DM, BM Gorman (1989). Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virol. 170: 561 – 565.
http://dx.doi.org/10.1016/0042-6822(89)90447-9
Mertens PPC, Brown F, Sanger DV (1984). Assignment of the genome segments of bluetongue virus type 1 to the proteins which they encode. Virol. 135: 207 – 217.
http://dx.doi.org/10.1016/0042-6822(84)90131-4
Prasad G, Minakshi, Malik Y (2007). 'Bluetongue' (Eds: G. Prasad, Minakshi and Yashpal Malik), A book published by Indian Council of Agricultural Research, New Delhi.
Ratinier M, Caporale M, Golder M, Franzoni G, Allan K, Nunes SF, Armezzani A, Bayoumy A, Rixon F, Shaw A, Palmarini M (2011). Identification and Characterization of a Novel Non–Structural Protein of Bluetongue Virus. PLoS Pathog 7(12): e1002477. doi:10.1371/journal.ppat.1002477.
http://dx.doi.org/10.1371/journal.ppat.1002477
Roy P (1989). Bluetongue virus genetics and genome structures. Review article. Virus Res. 13: 179 – 206.
http://dx.doi.org/10.1016/0168-1702(89)90015-4
Roy P, Marshall JA, French TJ (1990). Structures of the bluetongue virus genome and its encoded proteins. Curr. Top. Microbiol. Immunol. 162: 43 – 88.
http://dx.doi.org/10.1007/978-3-642-75247-6
http://dx.doi.org/10.1007/978-3-642-75247-6_3
PMid:2166648
Singh KP, Maan S, Samuel AR, Rao S, Meyer AJ, Mertens PPC (2004). Phylogenetic analysis of bluetongue virus genome segment 6 (encoding VP5) from different serotypes. Vet. Ital. 40: 479 – 483.
PMid:20422573
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 28: 2731 – 2739.
http://dx.doi.org/10.1093/molbev/msr121
PMid:21546353 PMCid:PMC3203626
Zhang Z, Schwartz S, Wagner L, Miller W (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7(1–2): 203 – 214.
http://dx.doi.org/10.1089/10665270050081478
PMid:10890397
Belhouchet M, MohdJaafar F, Firth AE, Grimes JM, Mertens PPC (2011) Detection of a fourth Orbivirus non–structural protein. PLoS ONE 6: e25697.
http://dx.doi.org/10.1371/journal.pone.0025697
PMid:22022432 PMCid:PMC3192121
Rao PP, Reddy YV, Meena K, Karunasree N, Susmitha, B, Uma M, Prasad PUVS, Chaitanya P, Reddy YN, Hegde NR (2012). Genetic characterization of bluetongue virus serotype 9 isolates from India. Virus Genes 44: 286 – 294.
http://dx.doi.org/10.1007/s11262-011-0707-4
PMid:22258368