Deciphering Foot and Mouth Disease Predictive Modeling: Uncovering Attribute Correlations and Risk Factors with Advanced Machine Learning
Deciphering Foot and Mouth Disease Predictive Modeling: Uncovering Attribute Correlations and Risk Factors with Advanced Machine Learning
Mokammel Hossain Tito*, Most Hoor E Jannat, Marzia Afrose, S.M. Jubayer Ahmed, Shah Md Maruf, Md. Arafat Hossain, Safiullah Samani, Ruksana Jahan Mira, Barshon Saha, Asraful Islam Jihad and Tonmoy Kumar Das
Bangabandhu Sheikh Mujibur Rahman Science and Technology University, Gopalganj, Bangladesh.
Abstract | Foot and mouth disease (FMD) presents a formidable challenge to the global livestock industry, with significant implications for food security, trade, and animal welfare. Despite advancements, FMD remains endemic, with a profound impact on economies worldwide. This study, conducted in Ethiopia’s East Wollega zone, we utilized a dataset comprising 266 bovine sera samples collected from Diga, Guto Gida, and Nekemte districts, employed machine learning (ML) algorithms to predict FMD outbreaks and assess attribute correlations. The dataset is taken from mendely with prevalence ranges from 4.8% to 72.1% in cattle. In this paper we have used total of five algorithms including Naïve Bayes, MLP (Multilayer Perceptron), SMO (Sequential Minimal Optimization), AdaBoostM1, and REP Tree. Each model evaluated using various criteria, such as Accuracy, Kappa statistic, Precision, Recall, F measure, Matthews Correlation Coefficient (MCC) and required time to perform the model. Analysis revealed Multilayer Perceptron (MLP) as the most effective model based on various evaluation criteria, achieving an impressive accuracy of 82.21%. Attributes were ranked by importance, with Age, Body Condition, and Physiology identified as the top critical factors. Moreover, eight association rules were derived, shedding light on attribute correlations in FMD occurrence. Our findings underscore the potential of ML in disease prediction and contribute valuable insights for proactive disease management strategies, offering a pathway towards safeguarding livestock and ensuring sustainable agricultural practices.
Editor | Muhammad Abubakar, National Veterinary Laboratories, Park Road, Islamabad, Pakistan.
Received | May 31, 2024; Accepted | August 06, 2024; Published | Septebmer 17, 2024
*Correspondence | Mokammel Hossain Tito, Bangabandhu Sheikh Mujibur Rahman Science and Technology University, Gopalganj, Bangladesh; Email: mokammel.17asvm014@bsmrstu.edu.bd
Citation | Tito, M.H., M.H.E. Jannat, M. Afrose, S.M.J. Ahmed, S.M. Maruf, M.A. Hossain, S. Samani, R.J. Mira, B. Saha, A.I. Jihad and T.K. Das. 2024. Deciphering foot and mouth disease predictive modeling: Uncovering attribute correlations and risk factors with advanced machine learning. Veterinary Sciences: Research and Reviews, 10(2): 58-71.
DOI | https://dx.doi.org/10.17582/journal.vsrr/2024/10.2.58.71
Keywords | FMD prediction, Mutilayer perceptron in FMD, Risk factor ranking, Association rules, Disease prevention
Copyright: 2024 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Foot and mouth disease (FMD) stands as a significant threat to the global livestock industry, with profound implications for food security, international trade, and animal welfare. This highly contagious viral infection affects cloven-hoofed animals, including cattle, sheep, pigs, and goats, leading to severe economic losses and societal disruption (Lewis et al., 2023; Seyoum and Tora, 2023). Despite considerable advancements in veterinary science and disease control measures, FMD remains endemic in many regions, posing ongoing challenges for animal health professionals, policymakers, and stakeholders (Aslam and Alkheraije, 2023; Jamal and Belsham, 2013).
FMD is caused by the foot-and-mouth disease virus (FMDV), a member of the Picornaviridae family, genus Aphthovirus. The virus exists in seven distinct serotypes, namely A, O, C, SAT1, SAT2, SAT3, and Asia-1, each exhibiting unique antigenic properties (Arzt et al., 2011). These serotypes display a varying geographical distribution, with certain strains predominating in specific regions. FMDV spreads rapidly through direct contact with infected animals, contaminated materials, or aerosolized droplets, facilitating transmission within and between livestock populations, prevalence ranges from 4.8% to 72.1% in cattle (Capozzo et al., 2021; Souley Kouato et al., 2018).
The clinical manifestations of FMD vary depending on the species affected and the virulence of the viral strain. Affected animals typically exhibit fever, vesicular lesions on the oral mucosa, hooves, and teats, accompanied by lameness, decreased appetite, and reduced milk production (Jamal and Belsham, 2013). Different study suggested that most cases resolve within weeks, severe outbreaks can lead to high morbidity and mortality rates, particularly in young or immunocompromised animals. Moreover, subclinical infections may occur, complicating disease surveillance and control efforts (Aslam and Alkheraije, 2023; Kompas et al., 2015).
Upon exposure, FMDV enters the host organism through mucous membranes or breaks in the skin, initiating viral replication in epithelial cells of the oral and nasal cavities. The virus then disseminates to regional lymph nodes, where it replicates further and enters the bloodstream, leading to systemic infection (Rodríguez-Habibe et al., 2020; Wang et al., 2015). The host immune response plays a critical role in disease outcome, with early production of neutralizing antibodies facilitating viral clearance and recovery. However, FMDV can evade host defenses through antigenic variation and immunomodulatory mechanisms, contributing to viral persistence and recurrence (Grubman et al., 2008; He et al., 2021)
The globalization of trade and the interconnectedness of global supply chains have heightened the risk of FMD spread across borders (Pattnaik et al., 2012). International movement of livestock, animal products, and vectors increases the likelihood of introducing exotic FMDV strains into susceptible populations, amplifying the threat of transboundary transmission (Sebhatu, 2019). Consequently, FMD outbreaks can have far-reaching consequences for international trade, leading to market closures, trade embargoes, and disruptions in global food supply chains. Mitigating these risks requires coordinated efforts at the national, regional, and international levels, encompassing surveillance, biosecurity measures, and contingency planning (WOAH).
Despite decades of research, several challenges persist in the understanding and management of FMD. These include the development of more effective vaccines and diagnostics, establishment of disease prediction strategy, elucidation of host-pathogen interactions, and enhancement of disease surveillance and control strategies (Capozzo et al., 2023; Santos et al., 2017).
Disease Prediction is a system that is based on machine learning (ML) and relies largely on user-provided symptoms to disease (Lowie et al., 2021; Romero et al., 2021). The most widely used model for predicting livestock disease outbreaks is based on classical statistical and mathematical techniques (Kouato et al., 2018). ML is a data science discipline that incorporates artificial intelligence and data science working with the algorithm that helps computers (the machine) learn from data and provide prediction outcomes (Grubman et al., 2008; Wang et al., 2020). The application of ML in animal health surveillance is beneficial, particularly in creating models that generate predictions (Punyapornwithaya et al., 2022; Tito et al., 2023). For instance, farms that are more inclined to become infected with a particular pathogen can be recognized. This is determined by analyzing previous case data and a set of potential risk factors (Ezanno et al., 2021; Tito et al., 2023). The application of machine learning has the potential to decipher methods for host susceptibility to disease (Uddin et al., 2019). The use of ML is plausible because dealing with large, complex, and hidden patterns can be daunting using other approaches (Wardeh et al., 2021). Moreover, data sequencing is becoming easier to do compared to decades ago and the cost reduction is largely due to this in the veterinary field (Munck et al., 2020). High accuracy in prediction can be achieved through the use of ML techniques (Uddin et al., 2019). Nowadays ML is being used in the field of human health disease identification including cardiac disease (Tito et al., 2024), Kidney disease (Bai et al., 2022), Lung disease (Harinath-Reddy et al., 2022), Skin disease (Verma et al., 2020), Diabetes (Hasan et al., 2020), Eye disease (Malik et al., 2019) and also used in animal disease including brucellosis (Tito, et al., 2023), vector borne disease (Tito et al., 2024), lumpy skin disease (Afshari Safavi, 2022). Now ML is also used in drug discovery (Vamathevan et al., 2019).
There has been a lack of research on developing prediction models to predict FMD outbreaks and risk factors analysis. The objectives of this paper are the identification of best ML model for the prediction of FMD, identification of the most important factors and association of the factors which are correlated for occurring the disease.
Data collection and processing
A cross-sectional study was undertaken to determine the seroprevalence of foot and mouth disease (FMD), ascertain the circulating serotypes of FMD virus, and evaluate potential risk factors associated with the disease occurrence in cattle within the Diga, Guto Gida, and Nekemte districts of the East Wollega zone, Ethiopia. The selection of study areas, districts, towns, and kebeles was performed purposively based on varying agro-ecologies and the presence of significant cattle populations.
Data collection
Data collection encompassed two main methodologies: questionnaire surveys and laboratory testing of a total of 266 bovine sera samples were collected from study animals utilizing a simple random sampling technique.
Data pre-processing
All the data are taken in an excel sheet. Missing values are identified and corrected using useful manner. Outliers and extreme values are removed from the dataset using WEKA.
Laboratory testing
The detection of nonstructural protein (NSP) antibodies of the Foot and Mouth Disease virus (FMDV) was conducted using the 3ABC ELISA method. Seroprevalence estimation was derived from these results. Additionally, specific serotypes of FMDV circulating in the study areas were identified utilizing Solid-phase competition ELISA (Borena et al., 2023).
Balancing of dataset
For the application of machine learning, balancing of dataset is important to predict the upcoming occurrence. Class imbalance is a common challenge in predictive modeling, especially in disease prediction tasks where the occurrence of diseased cases may be substantially lower than non-diseased ones. In our study, addressing this class imbalance is crucial for ensuring the robustness and accuracy of our predictive models.
To mitigate the effects of class imbalance, we employed SMOTE (Synthetic Minority Over-sampling Technique). SMOTE is a powerful method used to generate synthetic samples of the minority class by interpolating between existing minority class instances. By oversampling the minority class in a controlled manner, SMOTE helps to balance the class distribution within the dataset, thereby preventing the model from being biased towards the majority class (Blagus and Lusa, 2013).
The importance of SMOTE in our study lies in its ability to improve the generalization performance of our predictive models by providing them with a more representative training dataset (Li et al., 2014). Without addressing class imbalance, predictive models may exhibit poor performance, as they tend to favor the majority class due to its higher representation in the data (Dablain et al., 2023). By leveraging SMOTE, we aim to enhance the sensitivity of our models towards detecting diseased cases, ultimately leading to more accurate disease prediction outcomes. A total of 428 datasets were utilized, all devoid of missing values. In Figure 1, the blue bar denotes disease-positive instances, while the red bar signifies disease-negative instances. A noticeable imbalance between the two bars is evident upon closer inspection. This issue was addressed in Figure 2 through the application of SMOTE, achieving a balanced representation. The operational workflow depicted in Figure 3 outlines the comprehensive procedure detailed in the paper.
Modeling and evaluation metrics
In this study, five machine learning algorithms were selected to construct classifiers for classification tasks. We conducted a comprehensive evaluation of various classification models to identify the algorithm demonstrating superior performance across multiple metrics: Accuracy, Kappa Statistic (KS), Precision, Recall, F-measure, and computational Time. These metrics collectively gauge the models’ prediction quality, with detailed descriptions provided below:
Accuracy is determined by the ratio of correct predictions to the total number of predictions made as:

Where TP, FP, TN, FN represent true positive, false positive, true negative, and false negative, respectively.
The Kappa statistic is a robust metric for assessing inter-rater agreement for categorical data, offering a singular value that quantifies the extent of agreement beyond what is expected by chance. This measure is especially valuable in evaluating the performance of classifiers by examining the concordance between predicted and actual classifications.
Precision quantifies the accuracy of positive predictions by evaluating the proportion of true positives among all positive predictions made by the classifier. It serves as an indicator of the classifier’s capability to correctly identify instances that belong to the positive class.
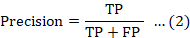
Recall, also referred to as True Positive Rate (TPR) or Sensitivity, quantifies the percentage of actual positive instances that are accurately detected. It is defined as the ratio of true positives to the total number of true positives and false negatives, and is mathematically expressed as:
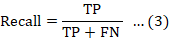
The F-measure, often referred to as the F1 score, represents the harmonic mean of precision and recall. This metric provides a balanced assessment of both precision and recall, making it particularly valuable in scenarios where false positives and false negatives hold equal significance.
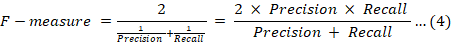
The Matthews correlation coefficient (MCC) is a robust metric for evaluating the quality of binary classifications. By incorporating true positives, true negatives, false positives, and false negatives, MCC offers a comprehensive and balanced measure of a classifier’s performance. This is particularly advantageous in scenarios involving imbalanced datasets, where traditional metrics may fall short.
The time required to construct a model is contingent upon various factors, including the size of the dataset, the complexity of the problem, and the selection of algorithms. This process generally involves iterative phases of data preprocessing, feature selection, and model training, all aimed at attaining optimal performance.
Machine learning classifiers description
Naïve Bayes, MLP (Multilayer Perceptron), SMO (Sequential Minimal Optimization), AdaBoostM1, and REP Tree are commonly utilized in disease prediction due to their diverse strengths in handling various aspects of the predictive modeling process including LSD (Afshari-Safavi, 2022), Brucellosis (Tito et al., 2023), Kindey disease (Almansour et al., 2019; Devika et al., 2019), Diabetes (AlThunayan et al., 2017) etc. Naïve Bayes, known for its simplicity and efficiency, is particularly adept at handling large datasets and is robust to noise, making it suitable for scenarios where computational resources are limited. MLP, a type of artificial neural network, excels at capturing complex nonlinear relationships in data, enabling it to uncover subtle patterns and nuances that may be indicative of disease susceptibility or progression. SMO, on the other hand, specializes in training support vector machines (SVMs), offering high accuracy and scalability, which are essential for dealing with large-scale datasets commonly encountered in disease prediction studies. AdaBoostM1 employs ensemble learning techniques, combining multiple weak learners to form a strong predictive model, thus enhancing the overall performance and robustness of the classifier. Lastly, REP Tree constructs decision trees using a rule-based approach, providing interpretable models that facilitate the identification of important risk factors and disease correlates. Together, these machine learning algorithms offer a comprehensive toolkit for disease prediction. Here we provided more clarification of each model:
Naïve bayes
Naïve Bayes employs a probabilistic approach to classify instances based on symptom patterns and risk factors associated with the disease. It assumes independence between predictors, simplifying calculations. By analyzing historical data on symptoms, patient characteristics, and disease outcomes, Naïve Bayes calculates the likelihood of a patient having the disease given their symptoms (Chen et al., 2020). It assigns the class label with the highest probability as the predicted disease status for the patient. This method enables early detection and intervention, enhancing patient outcomes and disease management. Naïve Bayes’s simplicity and efficiency make it a valuable tool in disease prediction and surveillance systems (Chen et al., 2020; Zhang and Li, 2007).
Multilayer perceptron
Multilayer Perceptron (MLP) algorithm utilizes artificial neural networks to model complex relationships between input variables (Figure 4) and disease occurrence. It consists of multiple layers of interconnected nodes, allowing it to capture intricate patterns in the data (Djerioui et al., 2020). MLP employs forward and backward propagation techniques to adjust weights and biases iteratively, optimizing the model’s performance. By training on historical data encompassing factors like genetic predispositions, lifestyle choices, and environmental influences, MLP can forecast disease likelihood with high accuracy (Mantzaris et al., 2008). Its ability to handle non-linear relationships makes it suitable for capturing the multifaceted nature of diseases. MLP’s versatility and adaptability make it a powerful tool in medical research for aiding in early diagnosis and risk assessment of various diseases (Djerioui et al., 2020; Mantzaris et al., 2008).
Sequential minimal optimization
In disease prediction, the SMO (sequential minimal optimization) algorithm is pivotal for its efficacy in training support vector machines (SVMs). SMO tackles quadratic optimization problems by iteratively solving smaller sub-problems, ensuring efficient SVM training even with large datasets (Arulanthu and Perumal, 2019). By analyzing various patient attributes like medical history, genetic markers, and environmental factors, SMO constructs an optimal decision boundary to classify instances into disease-positive or disease-negative categories. Its ability to handle high-dimensional data (Figure 5) and nonlinear relationships makes SMO particularly suitable for complex disease prediction tasks. SMO’s accurate classification aids in early diagnosis and proactive interventions, contributing to improved patient outcomes and public health management (Shalini et al., 2021).
AdaBoostM1
AdaBoostM1 algorithm, a meta-algorithm, combines multiple weak learners into a strong classifier. It iteratively trains weak classifiers on the data, focusing more on instances that were misclassified in previous iterations (Yadav and Pal, 2021). By giving higher weights to misclassified instances, AdaBoostM1 prioritizes learning from difficult-to-classify instances, improving overall performance. In the context of disease prediction, AdaBoostM1 could analyze various features such as symptoms, patient history, and environmental factors to identify patterns associated with the disease. Its ability to adaptively adjust to complex data distributions makes it particularly useful for detecting subtle patterns indicative of disease onset, enhancing diagnostic accuracy and early intervention strategies (Rawat, 2022).
REP tree
REP Tree, in disease prediction, constructs a decision tree by recursively partitioning the data into subsets based on attribute values. It uses a rule-based approach, selecting the best attribute to split the data at each node (Palaiokostas, 2021). REP tree aims to minimize impurity or uncertainty within each subset, optimizing predictive accuracy. In disease prediction, REP Tree analyzes various factors such as symptoms, patient history, and environmental conditions to classify instances into different disease categories or risk levels. It provides interpretable rules for understanding disease patterns and identifying important predictive factors. With its robustness and transparency, REP Tree is well-suited for decision support systems in healthcare, aiding in early diagnosis and personalized treatment planning (Gambhir et al., 2019).
Results and Discussion
Best model identification
In this paper, we employed a total of five machine learning algorithms selected based on their prior utilization in disease detection. The models were assessed using various evaluation criteria outlined in Table 1. Among them, MLP demonstrated the most promising performance for predicting FMD disease.
Table 1: Comparison of algorithms using evaluation criteria to assess performance in disease prediction studies
|
Parameter |
Machine learning models |
||||
|
Naïve Bayes |
MLP |
SMO |
AdaBoostM1 |
REP Tree |
|
|
Accuracy |
81.3953 % |
82.2093% |
86.0465% |
83.7209% |
83.7209% |
|
Kappa statistic |
0.6265 |
0.7431 |
0.719 |
0.6721 |
0.6718 |
|
Precision |
0.820 |
0.882 |
0.890 |
0.865 |
0.877 |
|
Recall |
0.814 |
0.872 |
0.860 |
0.837 |
0.837 |
|
F measure |
0.813 |
0.871 |
0.857 |
0.833 |
0.832 |
|
MCC |
0.633 |
0.753 |
0.749 |
0.700 |
0.711 |
|
Required time (s) |
0.01 |
0.33 |
0.02 |
0.02 |
0.01 |
Risk factors ranking
To predict the occurrence of the disease, we employed various factors known to contribute to its onset. Our study ranked these factors in Table 2 according to their importance in causing FMD, with Age > Body Condition > Physiology > Agro Climate > Sex > Breed.
Table 2: Risk factors ranking according to their impact on FMD.
|
Attributes |
Significance Value |
|
Age |
0.2222 |
|
Body Condition |
0.1112 |
|
Physiology |
0.0888 |
|
Agro Climate |
0.0574 |
|
Sex |
0.0547 |
|
Breed |
0.0139 |
Association rules
Table 3 presents the findings of our study, uncovering the associations between attributes that significantly contribute to disease occurrence. We identified eight (8) key rules that reliably determine disease status, each accompanied by its corresponding confidence percentage.
The global status of Foot-and-Mouth Disease (FMD) remains a significant concern, with sporadic outbreaks reported worldwide despite efforts in vaccination and control measures. Persistent surveillance and coordinated international strategies are crucial for containment and eventual eradication of this economically devastating livestock disease (WOAH; FAO). Here, we tried to identify the preventive measures for the prevention of the disease. The Table 1 represents the performance metrics of five machine learning models - Naïve Bayes, MLP (Multilayer Perceptron), SMO (Sequential Minimal Optimization), AdaBoostM1, and REP Tree (Qiu et al., 2021; Lin et al., 2021; Yazdanbakhsh et al., 2017; Zhang et al., 2021; Punyapornwithaya et al., 2022). The effectiveness of each model is assessed using various criteria, such as accuracy (Figure 6), Kappa statistic (Figure 7), Precision (Figure 8), Recall (Figure 9), F measure (Figure 10), and Matthews Correlation Coefficient (MCC) (Figure 11). Additionally, the required execution time for each model is illustrated in Figure 12. Among these models, SMO achieves the highest accuracy of 86.0465% among all the used models, closely followed by MLP with 82.2093%. However, when considering other metrics like precision, recall, and F-measure, MLP consistently outperforms others with values of 0.882, 0.872, and 0.871, respectively. Additionally, MLP exhibits the highest MCC of 0.753, indicating its robustness in handling imbalanced datasets. Despite its slightly longer computation time (0.33 seconds) compared to others, MLP emerges as the best model overall due to its fairly efficient compared to other models, based on the performance across various evaluation metrics.
Table 3: Association of factors which are correlated for the disease status.
|
Rule No. |
Combinations of clinical factors |
Disease status |
Accuracy |
|
1 |
Age=Old + Body Condition=Poor + Agro Climate=Lowland + Breed=Local |
Positive |
97% |
|
2 |
Sex=Female + Body Condition=Poor + Agro Climate=Lowland + Breed=Local |
Positive |
93% |
|
3 |
Sex=Female + Age=Old + Breed=Local |
Positive |
92% |
|
4 |
Sex=Female + Age=Old + Body Condition=Poor + Agro Climate= Lowland + Breed=Local |
Positive |
99% |
|
5 |
Age=Old + Body condition=Poor + Physiology = Lactating + Sex= Female |
Positive |
100% |
|
6 |
Sex=Male + Age=Old + Body condition=Poor |
Positive |
100% |
|
7 |
Sex=Female + Body Condition=Poor + Physiology = Heifer |
Positive |
100% |
|
8 |
Breed=Local + Physiology=Bull + Sex=Male |
Negative |
100% |
Ranked attributes provide invaluable insights into the underlying patterns and relationships within the data, which focuses on the most informative features while potentially reducing computational complexity. Moreover, understanding attribute importance facilitates domain knowledge discovery and aids in the creation of more accurate and robust predictive models (Calker et al., 2005). In the Table 2 we ranked our dataset attributes. The ranked attributes number value reflects the importance of each attribute in a dataset. Weka assigns a numerical value to attributes based on their relevance to the target variable or predictive power. Higher values indicate greater influence in predicting the target, while lower values signify less contribution to predictive accuracy. This ranked list of attributes helps to identify and select the most informative features for their machine learning models, thereby improving model performance, reducing overfitting, and enhancing interpretability. Older animals tends to have susceptibility than others where poor body conditioned and heifer showed more affinity, local breed showed more disease than cross breed.
Weka association rules are crucial as they provide insights into complex relationships within datasets, aiding in decision-making across various domains. By uncovering patterns and associations, these rules facilitate predictive modeling, and decision support systems, enhancing both research and practical applications (Lekha et al., 2013). It is able to extract meaningful associations from large datasets makes them indispensable for data-driven research, offering valuable insights into intricate relationships and dependencies. Utilizing Weka association rules ensures rigorous analysis (Tanna and Ghodasara, 2014).
Table 3 presents the eight (8) most significant rules generated from the Apriori algorithm (association rules), demonstrating correlations among various attributes. Rule 1 indicates a 97% likelihood of FMD occurrence when age is old, body condition is poor, agro climate is lowland, and breed is local. Rule 2 exhibits a 93% confidence level in predicting disease positivity when the sex is female, body condition is poor, agro climate is lowland, and breed is local. The 3rd rule suggests a 92% possibility of disease occurrence with attributes including female sex, old age, and local breed. Rule 4 highlights a 99% likelihood of disease suspicion when female sex, old age, poor body condition, local breed, and lowland agro climate are present. Rules 5, 6, and 7 demonstrate the machine’s 100% confidence in predicting positive tests under various conditions involving old age, poor body condition, and specific physiological states or sexes. Conversely, Rule 8 indicates 100% confidence in predicting a negative outcome when attributes such as local breed, bull physiology, and male sex are observed. These rules elucidate the correlations among different attributes regarding disease occurrence or absence.
The REP tree Figure 13 illustrates a decision tree-based model for disease prediction, depicting hierarchical splits based on relevant attributes. Each node represents a decision point, with branches indicating different attribute values leading to classification outcomes. The tree structure offers interpretability, facilitating the identification of key risk factors and their influence on disease prediction. Circular node is the root node and rectangle represents the leaf node where decisions are made (Saha et al., 2022).
Limitations of using ML models for disease prediction
Data quality: ML model accuracy depends on high-quality data. Issues like incomplete records and inconsistent formats can harm performance.
Generalizability: Models may not perform well on new data due to demographic and environmental variations. Validation across diverse populations is essential.
Feature selection: Relevant features are crucial. Irrelevant features add noise, while missing important ones lead to underfitting.
Overfitting: Complex models may capture noise instead of the actual signal, performing well on training data but poorly on new data. Cross-validation and regularization help mitigate this.
Challenges in implementing proactive measures based on ML predictions
Data quality: Incomplete or inconsistent data can hinder reliable predictions. Ensuring data privacy is also challenging.
Model accuracy: False positives/negatives and poor generalizability across regions can affect reliability.
Implementation: Requires significant infrastructure and skilled personnel. Resistance to adopting new technologies may occur.
Ethical and regulatory issues: Ethical considerations and regulatory compliance are complex.
Cost: Developing and maintaining ML models is costly, and efficient resource allocation is necessary.
Conclusions and Recommendaions
In our investigation, Multilayer Perceptron (MLP) exhibited superior performance across multiple evaluation metrics from where new disease status can be predicted and which will be 82.2093% accurate. Additionally, our analysis revealed the ranking of factors in terms of their importance for Foot-and-Mouth Disease (FMD) where age is the most important factor then Body Condition then Physiology then Agro Climate then Sex and lastly Breed among all the factors. Furthermore, we have derived eight (8) rules delineating the correlations among attributes contributing to FMD occurrence. Important factors, correlation of the attributes of this study can improve disease prevention, benefiting farmers, farm owners and the disease can be controlled.
It is recommended to diversify dataset sources beyond Ethiopia. Conducting surveys across multiple regions globally will enable a comprehensive understanding of these factors. Pooling together findings from diverse geographical areas will facilitate more robust decision-making processes. Employing advanced analytical techniques to integrate and analyze heterogeneous datasets can enhance the depth and accuracy of insights gleaned from such comparative studies.
Acknowledgements
The authors express their gratitude to the Bangabandhu Sheikh Mujibur Rahman Science and Technology University Gopalganj, Bangladesh. Also, we extend our sincere appreciation to the Weka Team at the University of Waikato, New Zealand, for their invaluable assistance with data analysis.
Novelty Statement
Our model evaluation criteria differed from others, and our results were superior. Additionally, we ranked the attributes based on their significance and demonstrated the correlation between the factors responsible for the occurrence of the disease. It will be more reliable in the field of veterinary medicine.
Author’s Contribution
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Most Hoor E Jannat, Marzia Afrose, S. M. Jubayer Ahmed, Shah Md Maruf, Md. Arafat Hossain, Safiullah Samani, Ruksana Jahan Mira, Barshon Saha, Asraful Islam Jihad and Tonmoy Kumar Das. The first draft of the manuscript was written by Mokammel Hossain Tito and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available in the Mendeley Data repository, (https://data.mendeley.com/datasets/yr26nj97zb/1).
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit organizations.
Conflict of interest
The authors have declared no conflict of interest.
References
Afshari-Safavi, E., 2022. Assessing machine learning techniques in forecasting lumpy skin disease occurrence based on meteorological and geospatial features. Trop. Anim. Hlth. Prod., 54(1): 55. https://doi.org/10.1007/s11250-022-03073-2
Almansour, N.A., Syed, H.F., Khayat, N.R., Altheeb, R.K., Juri, R.E., Alhiyafi, J., Alrashed, S. and Olatunji, S.O., 2019. Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study. Comp. Biol. Med., 109: 101–111. https://doi.org/10.1016/j.compbiomed.2019.04.017
Al-Thunayan, L., Al-Sahdi, N. and Syed, L., 2017. Comparative analysis of different classification algorithms for prediction of diabetes disease. Proceedings of the second international conference on internet of things, data and cloud computing, pp. 1–6. https://doi.org/10.1145/3018896.3036387
Arulanthu, P. and Perumal, E., 2019. Predicting the chronic kidney disease using various classifiers. 2019 4th international conference on electrical, electronics, communication, computer technologies and optimization techniques (ICEECCOT). pp. 70–75. https://doi.org/10.1109/ICEECCOT46775.2019.9114653
Arzt, J., Juleff, N., Zhang, Z. and Rodriguez, L.L., 2011. The pathogenesis of foot-and-mouth disease I: Viral pathways in cattle. Transbound. Emerg. Dis., 58(4): 291–304. https://doi.org/10.1111/j.1865-1682.2011.01204.x
Aslam, M. and Alkheraije, K.A., 2023. The prevalence of foot-and-mouth disease in Asia. Front. Vet. Sci., 10. https://doi.org/10.3389/fvets.2023.1201578
Bai, Q., Su, C., Tang, W. and Li, Y., 2022. Machine learning to predict end stage kidney disease in chronic kidney disease. Sci. Rep., 12(1): 8377. https://doi.org/10.1038/s41598-022-12316-z
Blagus, R. and Lusa, L., 2013. SMOTE for high-dimensional class-imbalanced data. BMC Bioinf., 14(1): 106. https://doi.org/10.1186/1471-2105-14-106
Borena, B.M., Tolawak, D., Muluneh, A., Chibssa, T. and Mekonnen, G., 2023. Seroprevalence, serotype, and associated risk factors of foot and mouth disease in selected districts of East Wollega Zone, Ethiopia. 1. https://doi.org/10.2139/ssrn.4328193
Calker, K.J.V., Berentsen, P.B.M., Giesen, G.W.J. and Huirne, R.B.M., 2005. Identifying and ranking attributes that determine sustainability in Dutch dairy farming. Agric. Hum. Values, 22(1): 53–63. https://doi.org/10.1007/s10460-004-7230-3
Capozzo, A.V., Pérez-Filgueira, M., Vosloo, W. and Gay, C.G., 2021. Editorial: FMD research: Bridging the gaps with novel tools. Front. Vet. Sci., 8. https://doi.org/10.3389/fvets.2021.686141
Capozzo, A.V., Vosloo, W., de los Santos, T., Pérez, A.M. and Pérez-Filgueira, M., 2023. Editorial: Foot-and-mouth disease epidemiology, vaccines and vaccination: Moving forward. Front. Vet. Sci., 10. https://doi.org/10.3389/fvets.2023.1231005
Chen, S., Webb, G.I., Liu, L. and Ma, X., 2020. A novel selective naïve Bayes algorithm. Know. Based Syst., 192: 105361. https://doi.org/10.1016/j.knosys.2019.105361
Dablain, D., Krawczyk, B. and Chawla, N.V., 2023. DeepSMOTE: Fusing deep learning and SMOTE for imbalanced data. IEEE Trans. Neural Netw. Learn. Syst., 34(9): 6390–6404. https://doi.org/10.1109/TNNLS.2021.3136503
Devika, R., Avilala, S.V. and Subramaniyaswamy, V., 2019. Comparative study of classifier for chronic kidney disease prediction using Naive Bayes, KNN and random forest. 2019 3rd international conference on computing methodologies and communication (ICCMC), pp. 679–684. https://doi.org/10.1109/ICCMC.2019.8819654
Djerioui, M., Brik, Y., Ladjal, M. and Attallah, B., 2020. Heart Disease prediction using MLP and LSTM models. 2020 international conference on electrical engineering (ICEE): 1–5. https://doi.org/10.1109/ICEE49691.2020.9249935
Ezanno, P., Picault, S., Beaunée, G., Bailly, X., Muñoz, F., Duboz, R., Monod, H. and Guégan, J.F., 2021. Research perspectives on animal health in the era of artificial intelligence. Vet. Res., 52(1): 40. https://doi.org/10.1186/s13567-021-00902-4
Gambhir, S., Kumar, Y., Malik, S., Yadav, G. and Malik, A., 2019. Early diagnostics model for dengue disease using decision tree-based approaches. In Pre-screening systems for early disease prediction, detection, and prevention, IGI Global. pp. 69–87. https://doi.org/10.4018/978-1-5225-7131-5.ch003
Grubman, M.J., Moraes, M.P., Diaz-San Segundo, F., Pena, L. and De Los Santos, T., 2008. Evading the host immune response: How foot-and-mouth disease virus has become an effective pathogen. FEMS Immunol. Med. Microbiol., 53(1): 8–17. https://doi.org/10.1111/j.1574-695X.2008.00409.x
Harinath-Reddy, C., Koushik Kumar, B.V., Sai Teja Varma, N., Vidya, S., Nagaraj, P. and Muthamil Sudar, K., 2022. Risk prediction of lung disease using deep learning approach. In: Second international conference on image processing and capsule networks (eds. J.I.Z. Chen, J.M.R.S. Tavares, A.M. Iliyasu and K.L. Du). Springer International Publishing. pp. 462–471. https://doi.org/10.1007/978-3-030-84760-9_40
Hasan, M.K., Alam, M.A., Das, D., Hossain, E. and Hasan, M., 2020. Diabetes prediction using ensembling of different machine learning classifiers. IEEE Access, 8: 76516–76531. https://doi.org/10.1109/ACCESS.2020.2989857
He, Y., Li, K., Cao, Y., Sun, Z., Li, P., Bao, H., Wang, S., Zhu, G., Bai, X., Sun, P., Liu, X., Yang, C., Liu, Z., Lu, Z., Rao, Z. and Lou, Z., 2021. Structures of foot-and-mouth disease virus with neutralizing antibodies derived from recovered natural host reveal a mechanism for cross-serotype neutralization. PLOS Pathog., 17(4): e1009507. https://doi.org/10.1371/journal.ppat.1009507
Jamal, S.M. and Belsham, G.J., 2013. Foot-and-mouth disease: Past, present and future. Vet. Res., 44(1): 116. https://doi.org/10.1186/1297-9716-44-116
Kompas, T., Nguyen, H.T.M. and Ha, P.V., 2015. Food and biosecurity: Livestock production and towards a world free of foot-and-mouth disease. Food Secur., 7(2): 291–302. https://doi.org/10.1007/s12571-015-0436-y
Kouato, B.S., Clercq, K.D., Abatih, E., Pozzo, F.D., King, D.P., Thys, E., Marichatou, H. and Saegerman, C., 2018. Review of epidemiological risk models for foot-and-mouth disease: Implications for prevention strategies with a focus on Africa. PLoS One, 13(12): e0208296. https://doi.org/10.1371/journal.pone.0208296
Lekha, A., Srikrishna, C.V. and Vinod, V., 2013. Utility of association rule mining: A case study using Weka tool. 2013 International conference on emerging trends in VLSI, embedded system, nano electronics and telecommunication system (ICEVENT), pp. 1–6. https://doi.org/10.1109/ICEVENT.2013.6496554
Lewis, R.A., Kashongwe, O.B. and Bebe, B.O., 2023. Quantifying production losses associated with foot and mouth disease outbreaks on large-scale dairy farms in Rift valley, Kenya. Trop. Anim. Hlth. Prod., 55(5): 293. https://doi.org/10.1007/s11250-023-03707-z
Li, K., Zhang, W., Lu, Q. and Fang, X., 2014. An improved SMOTE imbalanced data classification method based on support degree. 2014 international conference on identification, information and knowledge in the internet of things, pp. 34–38. https://doi.org/10.1109/IIKI.2014.14
Lin, X., Wang, X., Wang, Y., Du, X., Jin, L., Wan, M., Ge, H. and Yang, X., 2021. Optimized neural network based on genetic algorithm to construct hand-foot-and-mouth disease prediction and early-warning model. Int. J. Environ. Res. Publ. Hlth., 18(6): Article 6. https://doi.org/10.3390/ijerph18062959
Lowie, T., Callens, J., Maris, J., Ribbens, S. and Pardon, B., 2021. Decision tree analysis for pathogen identification based on circumstantial factors in outbreaks of bovine respiratory disease in calves. Prevent. Vet. Med., 196: 105469. https://doi.org/10.1016/j.prevetmed.2021.105469
Malik, S., Kanwal, N., Asghar, M.N., Sadiq, M.A.A., Karamat, I. and Fleury, M., 2019. Data driven approach for eye disease classification with machine learning. Appl. Sci., 9(14): Article 14. https://doi.org/10.3390/app9142789
Mantzaris, D.H., Anastassopoulos, G.C. and Lymberopoulos, D.K., 2008. Medical disease prediction using artificial neural networks. 2008 8th IEEE Int. Conf. BioInf. BioEng., pp. 1–6. https://doi.org/10.1109/BIBE.2008.4696782
Munck, N., Njage, P.M.K., Leekitcharoenphon, P., Litrup, E. and Hald, T., 2020. Application of whole-genome sequences and machine learning in source attribution of salmonella typhimurium. Risk Anal., 40(9): 1693–1705. https://doi.org/10.1111/risa.13510
Palaiokostas, C., 2021. Predicting for disease resistance in aquaculture species using machine learning models. Aquacult. Rep., 20: 100660. https://doi.org/10.1016/j.aqrep.2021.100660
Pattnaik, B., Subramaniam, S., Sanyal, A., Mohapatra, J.K., Dash, B.B., Ranjan, R. and Rout, M., 2012. Foot-and-mouth disease: Global status and future road map for control and prevention in India. Agric. Res., 1(2): 132–147. https://doi.org/10.1007/s40003-012-0012-z
Punyapornwithaya, V., Klaharn, K., Arjkumpa, O. and Sansamur, C., 2022. Exploring the predictive capability of machine learning models in identifying foot and mouth disease outbreak occurrences in cattle farms in an endemic setting of Thailand. Prevent. Vet. Med., 207: 105706. https://doi.org/10.1016/j.prevetmed.2022.105706
Qiu, J., Qiu, T., Dong, Q., Xu, D., Wang, X., Zhang, Q., Pan, J. and Liu, Q., 2021. Predicting the antigenic relationship of foot-and-mouth disease virus for vaccine selection through a computational model. IEEE/ACM Trans. Comp. Biol. Bioinf., 18(2): 677–685. https://doi.org/10.1109/TCBB.2019.2923396
Rawat, D., 2022. Validating and strengthen the prediction performance using machine learning models and operational research for lung cancer. 2022 IEEE international conference on data science and information system (ICDSIS), pp. 1–5. https://doi.org/10.1109/ICDSIS55133.2022.9915898
Rodríguez-Habibe, I., Celis-Giraldo, C., Patarroyo, M.E., Avendaño, C. and Patarroyo, M.A., 2020. A comprehensive review of the immunological response against foot-and-mouth disease virus infection and its evasion mechanisms. Vaccines, 8(4): Article 4. https://doi.org/10.3390/vaccines8040764
Romero, M.P., Chang, Y.M., Brunton, L.A., Prosser, A., Upton, P., Rees, E., Tearne, O., Arnold, M., Stevens, K. and Drewe, J.A., 2021. A comparison of the value of two machine learning predictive models to support bovine tuberculosis disease control in England. Prevent. Vet. Med., 188: 105264. https://doi.org/10.1016/j.prevetmed.2021.105264
Saha, S., Sarkar, R., Roy, J., Saha, T.K., Bhardwaj, D. and Acharya, S., 2022. Predicting the landslide susceptibility using ensembles of bagging with RF and REP Tree in Logchina, Bhutan. In: Impact of climate change, land use and land cover, and socio-economic dynamics on landslides (eds. R. Sarkar, R. Shaw and B. Pradhan). Springer Nature. pp. 275–298. https://doi.org/10.1007/978-981-16-7314-6_12
Santos, D.V. dos, Silva, G.S.E, Weber, E.J., Hasenack, H., Groff, F.H.S., Todeschini, B., Borba, M.R., Medeiros, A.A.R., Leotti, V.B., Canal, C.W. and Corbellini, L.G., 2017. Identification of foot and mouth disease risk areas using a multi-criteria analysis approach. PLoS One, 12: e0178464. https://doi.org/10.1371/journal.pone.0178464
Sebhatu, T.T., 2019. Foot-and-mouth disease. In: Transboundary animal diseases in Sahelian Africa and connected regions (eds. M. Kardjadj, A. Diallo and R. Lancelot). Springer International Publishing. pp. 207–231. https://doi.org/10.1007/978-3-030-25385-1_11
Seyoum, W. and Tora, E., 2023. Foot and mouth disease in Ethiopia: A systematic review and meta-analysis in the last fifteen years (2007–2021). Bull. Natl. Res. Centre, 47(1): 32. https://doi.org/10.1186/s42269-023-01004-1
Shalini, Saini, P.K. and Sharma, Y.M., 2021. An intelligent hybrid model for forecasting of heart and diabetes diseases with SMO and ANN. In: Intelligent energy management technologies (eds. M.S. Uddin, A. Sharma, K.L. Agarwal and M. Saraswat). Springer. pp. 133–138. https://doi.org/10.1007/978-981-15-8820-4_13
Souley Kouato, B., De Clercq, K., Abatih, E., Dal Pozzo, F., King, D.P., Thys, E., Marichatou, H. and Saegerman, C., 2018. Review of epidemiological risk models for foot-and-mouth disease: Implications for prevention strategies with a focus on Africa. PLoS One, 13(12): e0208296. https://doi.org/10.1371/journal.pone.0208296
Tanna, P. and Ghodasara, Y., 2014. Using Apriori with WEKA for frequent pattern mining. Int. J. Eng. Trends Technol., 12(3): 127–131. https://doi.org/10.14445/22315381/IJETT-V12P223
Tito, M.H., Arifuzzaman, M., Jannat, M.H.E., Nasrin, A., Gazder, U., Asaduzzaman, M., Ashrafuzzaman, M. and Maruf, S.M., 2023. Revolutionizing brucellosis disease prediction with specialized machine learning techniques. 7th IET Smart Cities Symp., 2023: 388–393. https://doi.org/10.1049/icp.2024.0956
Tito, M.H., Arifuzzaman, M., Jannat, M.H.E., Rahman, M.S., Sharmy, S.T., Nasrin, A., Asaduzzaman, M., Ashrafuzzaman, M., Prince, D.B. and Asif, A.H., 2023. A comparative study of ensemble machine learning algorithms for brucellosis disease prediction: Detection of brucellosis using artificial intelligence. Lett. Anim. Biol., 3(2): Article 2. https://doi.org/10.62310/liab.v3i2.119
Tito, M.H., Arifuzzaman, M., Nasrin, A., Khan, S., Asaduzzaman, M., Chohan, M.S. and Al-Duais, A.N., 2024. Deep learning for prediction of cardiovascular disease. 2024 ASU international conference in emerging technologies for sustainability and intelligent systems (ICETSIS), pp. 599–603. https://doi.org/10.1109/ICETSIS61505.2024.10459447
Tito, M.H., Arifuzzaman, M., Rahman, M.S., Khan, S., Chohan, M.S. and Nasrin, A., 2024. Predictive modeling of global vector-borne diseases: Leveraging machine learning for intervention strategies. 2024 ASU international conference in emerging technologies for sustainability and intelligent systems (ICETSIS): pp. 1027–1031. https://doi.org/10.1109/ICETSIS61505.2024.10459646
Tito, M.H., Rahman, M.S., Jannat, M.H.E., Sharmy, S.T., Nasrin, A. and Asaduzzaman, M., 2023. Prediction of brucellosis disease with ensemble machine learning application. 7th IET smart cities symposium (SCS 2023), 2023: 163–166. https://doi.org/10.1049/icp.2024.0918
Uddin, S., Khan, A., Hossain, M.E. and Moni, M.A., 2019. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inf. Decision Making, 19(1): 281. https://doi.org/10.1186/s12911-019-1004-8
Vamathevan, J., Clark, D., Czodrowski, P., Dunham, I., Ferran, E., Lee, G., Li, B., Madabhushi, A., Shah, P., Spitzer, M. and Zhao, S., 2019. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Dis., 18(6): 463–477. https://doi.org/10.1038/s41573-019-0024-5
Verma, A.K., Pal, S. and Kumar, S., 2020. Prediction of skin disease using ensemble data mining techniques and feature selection method. A comparative study. Appl. Biochem. Biotechnol., 190(2): 341–359. https://doi.org/10.1007/s12010-019-03093-z
Wang, G., Wang, Y., Shang, Y., Zhang, Z. and Liu, X., 2015. How foot-and-mouth disease virus receptor mediates foot-and-mouth disease virus infection. Virol. J., 12(1): 9. https://doi.org/10.1186/s12985-015-0246-z
Wang, W., Kiik, M., Peek, N., Curcin, V., Marshall, I.J., Rudd, A.G., Wang, Y., Douiri, A., Wolfe, C.D. and Bray, B., 2020. A systematic review of machine learning models for predicting outcomes of stroke with structured data. PLoS One, 15(6): e0234722. https://doi.org/10.1371/journal.pone.0234722
Wardeh, M., Blagrove, M.S.C., Sharkey, K.J. and Baylis, M., 2021. Divide-and-conquer: Machine-learning integrates mammalian and viral traits with network features to predict virus-mammal associations. Nat. Commun., 12(1): 3954. https://doi.org/10.1038/s41467-021-24085-w
Yadav, D.C. and Pal, S., 2021. An ensemble approach for classification and prediction of diabetes mellitus disease. In: Emerging trends in data driven computing and communications (R. Mathur, C.P. Gupta, V. Katewa, D.S. Jat and N. Yadav). Springer, pp. 225–235. https://doi.org/10.1007/978-981-16-3915-9_18
Yazdanbakhsh, O., Zhou, Y. and Dick, S., 2017. An intelligent system for livestock disease surveillance. Inf. Sci., 378: 26–47. https://doi.org/10.1016/j.ins.2016.10.026
Zhang, H. and Li, D., 2007. Naïve bayes text classifier. 2007 IEEE international conference on granular computing (GRC 2007): 708–708.
Zhang, S., Su, Q. and Chen, Q., 2021. Application of machine learning in animal disease analysis and prediction. Curr. Bioinf., 16(7): 972–982. https://doi.org/10.2174/1574893615999200728195613
To share on other social networks, click on any share button. What are these?






