Genetic diversity of Fusarium Isolated from Members of Sternorrhyncha (Hemiptera): Entomopathogens against Bemisia tabaci
Genetic diversity of Fusarium Isolated from Members of Sternorrhyncha (Hemiptera): Entomopathogens against Bemisia tabaci
Waheed Anwar*, Muhammad S. Haider, Ahmad A. Shahid, Hamid Mushtaq, Usman Hameed, Muhammad Zia Ur Rehman and Muhammad Javed Iqbal
Institute of Agricultural Sciences, University of the Punjab, 54590, Lahore, Pakistan.
ABSTRACT
In the present study, fungal flora from families Aleyrodidae, Aphididae and Coccidae of the order Hemiptera, suborder Sternorrhyncha, were isolated from three different agroecological zones of Pakistan. Fusarium equiseti, Fusarium solani, Fusarium incarnatum and Fusarium sp. along with other fungi were isolated and characterized morphologically as well as genetically by amplifying internal transcribed spacer region (ITS). Variability among Fusarium species based on ITS showed that it is not enough to score diversity within species and isolates. Additionally pathogenicity of isolated Fusarium species was evaluated against nymph and adult of Bemisia tabaci. Under controlled conditions different species of Fusarium restrained the growth of B. tabaci as compared with control.
Article Information
Received 07 May 2016
Revised 15 August 2016
Accepted 23 September 2016
Available online 30 March 2017
Authors’ Contributions
MSH and AAS conceived and designed the study. WA, HM and JI surveyed and collected the samples. WA isolated and identified the fungi. WA and MZR maintained Bemisia tabaci and analyzed the data. WA and UH performed bioassays and wrote the article.
Key words
Fungi associated with insect, ITS, Bemisia tabaci, Entomopathogens, Fungal diversity
DOI: http://dx.doi.org/10.17582/journal.pjz/2017.49.2.639.645
* Corresponding author: waheedanwar.iags@pu.edu.pk
0030-9923/2017/0002-0639 $ 9.00/0
Copyright 2017 Zoological Society of Pakistan
INTRODUCTION
The sweet potato whitefly, Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae) is a major pest of economically important crops worldwide (Xu et al., 2012). B. tabaci is a species complex composed of more than 35 morphologically indistinguishable cryptic species which exhibit a wide range of genetic variations and many species of this complex show a specific pattern of geographic distribution (Liu et al., 2012; Tay et al., 2012). Some members of this species complex are considered major pests for a wide range of agricultural crops causing damage directly through excessive sap feeding and, indirectly through transmission of plant pathogenic viruses that whitefly acquire when feeding on phloem sap. There are 288 species of begomoviruses (family Gemniviridae) which are known to be transmitted by B. tabaci (Brown et al., 2015).
Fusarium species are mainly soil borne fungi and frequently associated with plant roots as parasites causing serious diseases on many plants all over the world (Bockus et al., 2007). Fusarium related diseases include citrus decline, vascular wilts, crown rot and root rot, head blight on cereal grain and Bakanae disease on rice (Safdar et al., 2013; Saremi et al., 2007). On the other hand some of Fusarium species were reported as entomopathogens and may live as saprophyte on dead insects (Teetor-Barsch and Roberts, 1983; Sun, 2008). As entomopathogens, some Fusarium species have been reported to cause moderate to high levels of infection, principally against homopterous and dipterous insects. However, low to moderate levels of infections with these entomopathogens have been reported from insects of other orders (e.g., Coleoptera and Lepidoptera) (Teetor-Barsch and Roberts, 1983). Many species of entomopathogenic Fusarium may kill their host insects through the activity of toxins produced by penetrating hyphae (Gupta et al., 1991).
Here, we also observed the genetic diversity of Fusarium species isolated from insects on the bases of Internal transcribed spacer region (ITS). ITS genetic diversity has been reported within species and individuals from a diverse range of eukaryotes, including insects (Fairley et al., 2005; Li and Wilkerson, 2007), marine sponges (Vierna et al., 2010) and fungi (Smith et al., 2007; Horton, 2002). The ITS regions are located between the repeating array of nuclear 18S, 5.8S and 28S ribosomal RNA genes and have 100-200 copies/genome. These regions are rapidly evolving and thus have been routinely used in species-level phylogeny in a wide range of organisms (Blouin, 2002) and this region can be used for taxonomic evidence of fungi (Kolawole et al., 2015).
In the present study, different strains of Fusarium infecting families in the suborder Sternorrhyncha were isolated, identified on morphological and molecular bases. Further the pathogenicity of isolated Fusarium species were tested against B. tabaci. This study provides information about a potential use of fungus as biological control agent against whitefly.
MATERIALS AND METHODS
Collection of insect samples
Detailed survey of cotton fields for sample collection was conducted in different localities of three agroecological zones of Punjab, Pakistan. These regions include: (1) Hot arid Zone (Layyah) (2) Cotton Zone (Bahawalpur) (3) Central mixed zone (Lahore). Naturally dead Aphis gossypii from family Aphididae, whitefly from family Aleyrodidae and mealybug from family Coccidae from suborder Sternorrhyncha infected with fungi were collected from upper leaves from 3 to 4 feet above the soil level. Ten insect samples of each species from each field were collected, carried out separately in sterilized jars and stored at 4°C for isolation of total fungi associated with them.
Isolation of fungi
For the isolation of fungi, sabouraud dextrose peptone yeast extract agar (SDAY) plates with one quarter strength was used (1/4). Insect samples were surface sterilized with 1% sodium hypochloride solution for 1 min followed by washing with sterilized distilled water and transferred to SDAY media plates. Plates were incubated at 25±1°C for 7 days. Total isolated fungi from each insect were purified before being stored at 4°C as pure culture. Pure cultures were sub-cultured on SDAY media plates in order to conduct bioassay.
Characterization of fungi
Morphological characterizations of isolated fungi were studied using different dichotomous keys (John et al., 2006; Domsch et al., 1980; Seifert, 1996) and submitted to First Fungal Culture Bank of Pakistan (FCBP) for accessions numbers.
Identification based on morphological characterization was complimented with amplification and sequencing of internal transcribed spacer (ITS) region. For this purpose, total DNA of isolated fungi was extracted by modified CTAB method (Stenglein and Balatti, 2006). DNA was extracted by sub culturing fungi in SDAY broth media and it was incubated at 26 C for 7 days at 100 rpm. The extracted DNA was analyzed on 1% agarose gel. Internal transcribed spacer region was amplified using ITS1 and ITS4 primers (White et al., 1990). PCR amplification was carried out in 25 µL reaction volumes containing 20–100 ng genomic DNA, 1x PCR buffer, 1.5 mM MgCl2, 200 uM of each dNTP, 0.4 mM of each primer and 1 uL Taq polymerase. The PCR amplification reactions were carried under following conditions ; one cycle of denaturation for 3 min at 94°C followed by 30 cycles of denaturation for 1 min at 94°C, annealing at 50°C for 1 min, elongation at 72°C for 1 min and final extension of 72°C at 10 min. The amplified PCR products were resolved on 1% agarose gel. Amplified PCR products were purified using QIAquick PCR purification kit (Qiagen, Valencia, CA, United States). Purified PCR products were sequenced from 1st BASE Malaysia using above mentioned primer based on procedure described by Sanger et al. (1977). Sequences were submitted to the NCBI database for the assignment of Genbank accession numbers.
Sequence and statistical analysis
Alignment and determination of consensus sequences were carried out using BioEdit (Hall, 1999). Homologous sequences in the databases were searched using the Basic Local Alignment Search Tool (Altschul et al., 1997), while phylogenetic tree was constructed using MEGA software version 7.0. The statistical procedure for phylogenetic tree construction used was the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) while 10000 bootstrap replications were carried out.
Pathogenicity bioassay against Bemisia tabaci
The Fusarium isolates sub-cultured on Dextrose Peptone yeast extract Agar (SDAY) medium were utilized for preparation of stock solutions and tested for their pathogenicity against B. tabaci. The healthy nymph and adult B. tabaci were used for all screening bioassay procedure. The stock spore suspension of fungal concentration of 4.0 x 108 / mL was prepared with the help of haemocytometer and further diluted up to concentration of 4.0 x 104 /mL. Pathogenicity test was conducted on the test by spraying different spore suspension on healthy nymph and adult B. tabaci as compare to control treated with sterilized water.
Data regarding mortality rate in nymphs and adults of B. tabaci was observed after 24 h intervals by using modified Abbott’s formula (Fleming and Ratnakaran, 1985).
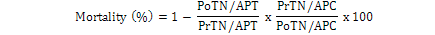
Where; PoTN, post-treated nymph; PrTN, pre-treated nymph; APT, adult population in treatment; APC, adult population in control.
Excel was used to compute the standard errors of means of five replicates. All the results were subjected to ANOVA followed by mean separation through Duncan’s multiple range test (P≤0.05) (Steel and Torrie, 1980) using computer software CO-STAT.
| FCBP acc. No. | Name of fungi | Source of isolation | Genbank No (ITS) |
| FCBP-EPF-1299 | Fusarium equiseti |
Aphis gossypii of Cotton [Field, IAGS], Central Mixed Zone |
LN827599 |
| FCBP-EPF-1301 | Fusarium incarnatum |
Bemisia tabaci, [IAGS field], Central Mixed Zone |
LN827601 |
| FCBP-EPF-1302 | Fusarium equiseti | Mealybug, [IAGS field], Central Mixed Zone | LN827600 |
| FCBP-EPF-1304 | Fusarium oxysporum |
Aphis gossypii, cotton field, [Bahawalpur], Cotton Zone |
N/A |
| FCBP-EPF-1376 |
Fusarium sp. |
Mealybug of cotton field, IAGS, [Lahore], Central Mixed Zone | LN827602 |
| FCBP-EPF-1389 | Fusarium equiseti |
Bemisia tabaci of cotton Field, IAGS, Central Mixed Zone |
LN827603 |
| FCBP-EPF-1420 | Fusarium solani |
Bemisia tabaci, [IAGS field], Central Mixed Zone |
LT159846 |
RESULTS AND DISCUSSION
Different fungi from various genera were isolated from different agroecological zones along with five species of Fusarium from families Aleyrodidae, Aphididae and Coccidae of order Hemiptera and suborder Sternorrhyncha,. Different species of Fusarium were isolated from Central mixed zone, Cotton zone and hot arid zone with many isolates. Details of Fusarium isolates are given in (Table I).
Genetic variations among isolates of fusarium species
Genetic variations among internal transcribed spacer region of Fusarium species isolated in this study were compared with the other isolates taken from NCBI. A phylogenetic tree was constructed and it was observed that Fusarium equiseti (LN827600) isolated from insect could not be categorized in the same group in which 2 other isolates (LN827599 and LN827603) formed a cluster with other isolates of F. equiseti taken from data base. Fusarium sp. (LN827602) and F. incarnatum (LN827601) both isolates of insects were classified in the same main cluster and having different sub clusters as mentioned in (Fig. 1). Whereas, F. solani (Sample X) isolated from insect was placed in the same cluster along with the other isolates of data base.
Pathogenicity bioassay against Bemisia tabaci
Pathogenicity of different isolated Fusarium species and their isolates were tested upon healthy nymph and adult whitefly to check mortality. It was observed that no isolate showed mortality till 48 h when 4.0 x 10 4 /mL spore suspension of different isolates of Fusarium were applied on nymph stage of B. tabaci. After 72 h, F. equiseti FCBP-EPF-1299 and FCBP-EPF-1389 showed maximum mortality of 26.67% while F. incarnatum FCBP-EPF-1301 showed minimum mortality of 13.33%.
Similarly after 96 and 120 h, maximum mortality was observed in isolates of F. equiseti followed by F. solani and F. oxysporum while minimum mortality was observed for F. incarnatum. After 144 h, all the isolates showed 100% mortality (Fig. 2A). Infection of Fusarium on nymph and adult B. tabaci were shown in Figure 4. When 4.0x108 / mL spore suspension of different isolates of Fusarium were applied on nymph stage of B. tabaci, it was observed that except F. incarnatum and F. oxysporum all isolates showed mortality after 48 h. After 72, 96 and 120 h, maximum mortality was observed in isolates of F. equiseti followed by F. solani and F. oxysporum while minimum mortality was noticed by F. incarnatum. After 144 h, all isolates of Fusarium showed 100% mortality (Fig. 2B).
A significant difference was observed in mortality rate of B. tabaci Nymph by isolates of F. equiseti as compared with other species. After different time durations a
significant difference was also observed by single isolate. When 4.0x104 /mL spore suspension of different isolates of Fusarium were applied on adult B. tabaci, it was observed that only F. oxysporum, Fusarium sp. and F. equiseti (FCBP-EPF-1302) showed 6.67% mortality up till 120 h. After 144 h, F. equiseti (FCBP-EPF-1302) and F. oxysporum showed 13.33% mortality while F. equiseti (FCBP-EPF-1389) and Fusarium sp. showed 6.67% mortality only (Fig. 3A).
When 4.0 x 10 8 / mL spore suspension of different Fusarium isolates were applied on adult B. tabaci, only F. oxysporum showed mortality after 96 h. After 120 h, F. equiseti (FCBP-EPF-1302), F. oxysporum and Fusarium sp. showed mortality. Maximum mortality of 20% was shown by F. equiseti (FCBP-EPF-1302) and F. oxysporum after 144 h (Fig. 3B).
There was no significant difference in mortality of adult B. tabaci between F. equiseti (FCBP-EPF-1302) and F. oxysporum (FCBP-EPF-1304) while significant difference in both was found when compared with other isolates used in this study.
In this study, we reported different species of Fusarium isolated from mealybug, aphid and whitefly. Most of the species were isolated first time from these insects and these fungi show mortality against Bemisia tabaci. Similarly, Jouda et al. (2010) isolated Fusarium sacchari and Fusarium semitectum from aphid and also showed Pathogenicity against it. Pelizza et al. (2011) reported F. verticillioides from grasshoppers. Fusarium equiseti (Corda) Sacc. was reported as a fungal parasite of the Brinjal mealybug, Coccidohystrix insolita (Green), in 1982 (Gopinathan et al., 1982). F. incarnatum-equiseti complex were also reported from a brown soft scale (Fan et al., 2014). These all studies showed that Fusarium is entomopathogenic in nature and cause Pathogenicity against different insects. According to our knowledge, isolates Fusarium equiseti are reported first time in this study from aphid, mealybug and Bemisia tabaci and it is obvious from the present study that among all the Fusarium species, F. equiseti (FCBP-EPF-1304) and F. oxysporum FCBP-1304 were most virulent and showed significant rate of mortality on nymph and adult B. tabaci.
Further studies are required for best optimization of conidial suspension of these Fusarium species for their effective use in future.
CONCLUSION
In this study, it is found that different species of Fusarium isolated from insects showed similarity on ITS region with the same species isolated from different sources. Fusarium equiseti and Fusarium oxysporum showed maximum mortality as compare to others species against nymph and adult B. tabaci so these species can be used in conserved biological control strategies against B. tabaci. Therefore further studies are necessary to know the proper infection mechanism of Fusarium species against whitefly.
ACKNOWLEDGMENTS
This research was funded in part through the Pakistan-U.S. Cotton Productivity Enhancement Program USDA Agricultural Research Service Project No 58-6402-0-178F. We also acknowledged the team of First Fungal Culture Bank of Pakistan (FCBP), Institute of Agricultural Sciences (IAGS), University of the Punjab, Lahore, Pakistan to provide guidance in the identification of fungi.
Statement of conflict of interest
Authors have declared no conflict of interest.
REFERENCES
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res., 25: 3389-3402. https://doi.org/10.1093/nar/25.17.3389
Blouin, M.S., 2002. Molecular prospecting for cryptic species of nematodes: mitochondrial DNA versus internal transcribed spacer. Int. J Parasitol., 32: 527-531. https://doi.org/10.1016/S0020-7519(01)00357-5
Brown, J.K., Zerbini, F.M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J.C. and Malathi, V.G., 2015. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Archiv. Virol., 160: 1593-1619. https://doi.org/10.1007/s00705-015-2398-y
Fairley, T.L., Kilpatrick, C.W. and Conn, J.E., 2005. Intragenomic heterogeneity of internal transcribed spacer rDNA in neotropical malaria vector Anopheles aquasalis (Diptera: Culicidae). J. med. Ent., 42: 795-800. https://doi.org/10.1603/0022-2585(2005)042[0795:ihoits]2.0.co;2
Fan, J.H., Xie, Y.P., Xue, J.L., Xiong, Q., Jiang, W.J., Zhang, Y.J. and Ren, Z.M., 2014. The strain HEB01 of Fusarium sp., a new pathogen that infects brown soft scale. Annls Microbiol., 64: 333-341. https://doi.org/10.1007/s13213-013-0668-z
Gopinathan, P.V., Beevi, N. and Nair, M.R.G.K., 1982. Occurrence of Fusarium equiseti (Corda) Sacc. as a fungal parasite of brinjal mealy bug Coccidohystrix insolita (Green). Entomon, 7: 120-121.
Gupta, S., Krasnoff, S.B., Underwood, N.L., Renwick, J.A.A. and Roberts, D.W., 1991. Isolation of beauvericin as an insect toxin from Fusarium semitectum and Fusarium moniliforme var. subglutinans. Mycopathologia, 115: 185-189. https://doi.org/10.1007/BF00462223
Horton, T.R., 2002. Molecular approaches to ectomycorrhizal diversity studies: variation in ITS at a local scale. In: Diversity and integration in Mycorrhizas. Springer Netherlands. pp. 29-39. https://doi.org/10.1007/978-94-017-1284-2_4
Jouda, G., Monia, B.H.K. and Naima, B., 2010. First report of aphidopathogenic fungi Fusarium semitectum (Berkeley and Ravenel, 1875) and Fusarium sacchari (Butler and Hafiz Khan) Gams (1971) on Capitophorus elaeagni (Del Guercio)(Hemiptera: Aphididae). Afr. J. Agric. Res., 5: 290-293.
Seifert, K., 1996. Fuskey: Fusarium Interactive Key (No. 632.4/S459). Agriculture and Agri-Food, Canada.
Kolawole, R.M., Thomas, B.T., Adekunle, A.A and Oluwadun, A., 2015. Postharvest pathogenic fungi of wheat circulating in Lagos State, Nigeria. Am. J. Res. Commun., 1: 421-428.
Li, C. and Wilkerson, R.C., 2007. Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae). J. Hered., 98: 51-59. https://doi.org/10.1093/jhered/esl037
Liu, B.M., Yan, F.M., Chu, D., Pan, H.P., Jiao, X.G., Xie, W., Wu, Q.J., Wang, S.L., Xu, B.Y., Zhou, X.G. and Zhang, Y.J., 2012. Difference in feeding behaviors of two invasive whiteflies on host plants with different suitability: implication for competitive displacement. Int. J. biol. Sci., 8: 697-706. https://doi.org/10.7150/ijbs.4108
Pelizza, S.A., Stenglein, S.A., Cabello, M.N., Dinolfo, M.I. and Lange, C.E., 2011. First record of Fusarium verticillioides as an entomopathogenic fungus of grasshoppers. J. Insect Sci., 11: 70. https://doi.org/10.1673/031.011.7001
Sanger, F., Nicklen, S. and Coulson, A.R., 1977. DNA sequencing with chain-terminating inhibitors. Proc. nat. Acad. Sci., 74: 5463-5467. https://doi.org/10.1073/pnas.74.12.5463
Saremi, H., Ammarellou, A. and Jafary, H., 2007. Incidence of crown rot disease of wheat caused by Fusarium pseudograminearum as a new soil born fungal species in North West Iran. Pakistan J. biol. Sci., 10: 3606-3612. https://doi.org/10.3923/pjbs.2007.3606.3612
Smith, M.E., Douhan, G.W. and Rizzo, D.M., 2007. Intra-specific and intra-sporocarp ITS variation of ectomycorrhizal fungi as assessed by rDNA sequencing of sporocarps and pooled ectomycorrhizal roots from a Quercus woodland. Mycorrhiza, 18: 15-22. https://doi.org/10.1007/s00572-007-0148-z
Stenglein, S.A. and Balatti, P.A., 2006. Genetic diversity of Phaeoisariopsis griseola in Argentina as revealed by pathogenic and molecular markers. Physiol. Mol. Pl. Pathol., 68: 158-167. https://doi.org/10.1016/j.pmpp.2006.10.001
Tay, W.T., Evans, G.A., Boykin, L.M. and De Barro, P.J., 2012. Will the real Bemisia tabaci please stand up? PLoS One, 7: e50550. https://doi.org/10.1371/journal.pone.0050550
Teetor-Barsch, G.H. and Roberts, D.W., 1983. Entomogenous Fusarium species. Mycopathologia, 84: 3-16. https://doi.org/10.1007/BF00436991
White, T.J., Bruns, T., Lee, S.J.W.T. and Taylor, J.W., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications, 18: 315-322.
Xu, C., Qiu, B., Cuthbertson, A.G., Zhang, Y. and Ren, S., 2012. Adaptability of sweetpotato whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) on seven marginal host plants. Int. J. Pest Manage., 58: 297-301. https://doi.org/10.1080/09670874.2012.678911
To share on other social networks, click on any share button. What are these?










