Genotyping Study for Infections of Theileria and Babesia in Cattle and Sheep in AL-Najaf Governorate Iraq
Maytham Askar Alwan Al-shabbani1, Hawraa Nasser rufaish1, Hassan Hachim Naser2, Murtadha Abbas3*, Hazem Almhanna4
1Department of Microbiology, Faculty of Veterinary Medicine, University of Kufa, Iraq; 2Department of Microbiology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Qadisiyah, Iraq; 3Department of public health, Faculty of Veterinary Medicine, University of Kufa, Iraq; 4Department of Anatomy, and Histology, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Qadisiyah, Iraq.
*Correspondence | Murtadha Abbas, Department of public health, Faculty of Veterinary Medicine. University of Kufa, Iraq; Email:
[email protected]
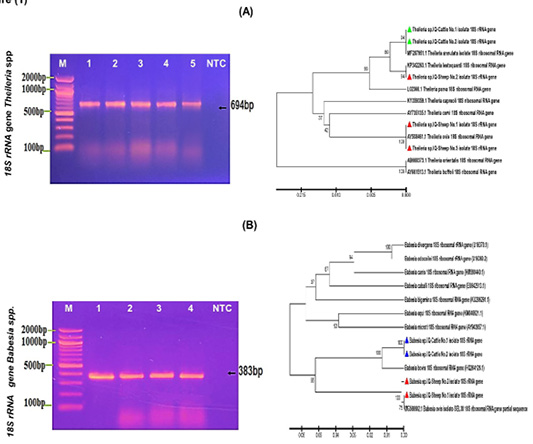
Figure 1:
(A) This figure illustrates the PCR product of the 18S rRNA gene in Theileria spp. of sheep and cattle blood samples at 694bp. M: DNA marker ladder (2000-100bp), and Lane (1-3) showed some positive samples of Theileria spp. from Sheep and Lane (4-5) positive samples of Theileria spp. from Cattle, and lane (NTC) Non-template negative control. Besides, the analysis of the phylogenetic tree of Theileria spp. is based on the 18S ribosomal rRNA gene partial sequence and genetic identification. The phylogenetic tree was constructed using the Unweighted Pair Group method with Arithmetic Mean (UPGMA tree) in the MEGA 6.0 version. Theileria spp. Sheep isolates (No. 1 and No. 3) were found to be closely related to NCBI-Blast Theileria ovis (AY508461.1), and Theileria spp. sheep isolates (No. 2) were identified to be closely related to NCBI-Blast Theileria lestoquardi (KP342263.1). Theileria spp. Cattle isolates (No. 1 and No. 2) were found to be closely related to NCBI-Blast Theileria annulata (MF287951.1) at a total genetic change (0.005-0.015%).(B): This figure illustrates the PCR product run on 1% Agarose gel electrophoresis and detected the 18S rRNA gene in Babesia spp. of sheep and cattle blood samples M: DNA marker ladder (2000-100bp), and Lane (1-2) showed some positive samples of Babesia spp. from Sheep and Lane (3-4) some positive samples of Babesia spp. from Cattle at 383bp PCR product size and lane (NTC) Non-template negative control. Besides, the analysis of the phylogenetic tree is based on 18S ribosomal rRNA gene partial sequence in the local Babesia species sheep and cattle isolates that were used for Babesia species genetic identification. The phylogenetic tree was analyzed using the Unweighted Pair Group method with Arithmetic Mean (UPGMA tree) in the MEGA 6.0 version. Babesia spp. sheep isolates (No.1 and No.2) were found to be closely related to NCBI-Blast Babesia ovis (MG569902.1) and Babesia spp. cattle isolates (No.1 and No. 2) were found to be closely related to NCBI-Blast Babesia bovis (HQ264126.1) at a total genetic change (0.01-0.06%).
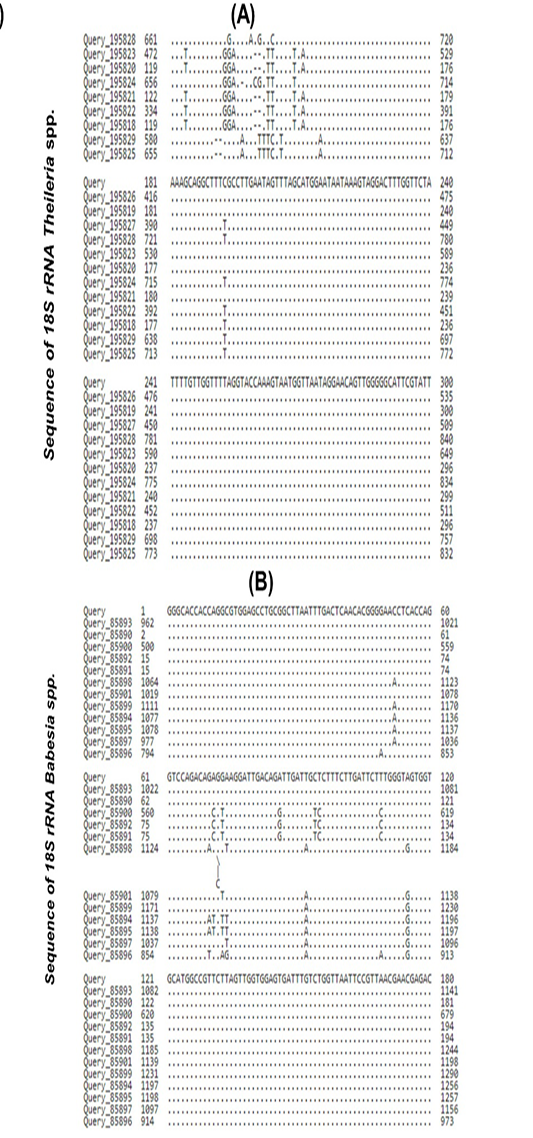
Figure 2:
(A) Multiple sequence alignment analysis for 18S ribosomal rRNA gene partial sequence in the local Theileria spp. isolates and the GenBank located Theileria spp. isolates based on the NCBI-BLAST Alignment tool. The figure shows similarity in the multiple alignment analysis (*) and mutation in the nucleotide sequences of the 18S ribosomal rRNA gene. (B): Multiple sequence alignment analysis for 18S ribosomal rRNA gene partial sequence in the local Babesia spp. isolates and the GenBank located Babesia spp. isolates based on the NCBI-BLAST Alignment tool. The figure shows similarity in the multiple alignment analysis (*) and mutation in the nucleotide sequences of the 18S ribosomal rRNA gene.