Lactic Acid Bacteria and Chickpea (Cicer arietinum L.): An Unexplored but Potentially Beneficial Liaison
Lactic Acid Bacteria and Chickpea (Cicer arietinum L.): An Unexplored but Potentially Beneficial Liaison
Ahmad Nadeem1,2 and Rubina Arshad1,2,*
1Nuclear Institute for Agriculture and Biology College (NIAB-C), Faisalabad
2Pakistan Institute of Engineering and Applied Sciences (PIEAS), Nilore, Islamabad
ABSTRACT
The present study was designed to explore plant-associated endophytic lactic acid bacteria (LAB), enhance their plant growth promoting efficacy by induced mutation and unlock their potential as bio-inoculant. Lactobacillus plantarum specific medium and non-selective media were used for isolation of LAB from plant cuttings. A total of seven isolates were isolated on the basis of colony morphology on De Man Rogosa Sharpe (MRS) agar plates, Gram staining, biochemical tests and molecular characterization. The co-culturing of LAB isolates with fungal cultures (in vitro) showed that among seven LAB isolates, only three possessed antifungal activity. One promising wild type isolate (LPA6) was subjected to gamma irradiation for mutation induction. Seeds of two chickpea varieties were inoculated with LAB isolates in four treatments comprising of T1 (wild type LPA6), T2 (mutant MLPA6), T3 (wild type LPA6 + mutant MLPA6 consortium) and T4 (un-inoculated control). Inoculated and uninoculated seeds were sown in net house in three replicates. In kabuli variety, maximum increase in plant height (16.7%), root length (5.9%), number of secondary branches (19%), pod number (63.2%) and seed number per plant (75%) was observed in MLPA6 mutant inoculated plants as compared to control whereas maximum increase in 100 seed weight (43.1%) and plant weight (90%) was attained in consortium treated plants over control. In desi variety, MLPA6 mutant also manifested significant increase in root length (53%), pod number (43.5%), seed number (50%), 100 seed weight (24%) and plant weight (67.9%) than control. These eco-friendly plant probiotics offer great potential for crop improvement and could further be exploited by conducting field trials for investigating their potential as an alternative to agrochemicals.
Article Information
Received 30 September 2020
Revised 12 December 2020
Accepted 10 February 2021
Available online 10 February 2022
(early access)
Published 22 October 2022
Authors’ Contribution
AN contributed to the overall execution of the study, analytical work, data collection and write-up of the first draft of manuscript. RA contributed to the conception and design of study, overall supervision during research work, statistical analysis of data and revision of manuscript.
Key words
Lactic acid bacteria, Endophytes, Plant probiotics, Mutation, Chickpea, Plant growth promotion, Biocontrol.
DOI: https://dx.doi.org/10.17582/journal.pjz/20200930120959
* Corresponding author: [email protected]
0030-9923/2023/0001-153 $ 9.00/0
Copyright 2023 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Lactic acid bacteria (LAB) are a group mainly characterized by being Gram-positive, rod or cocci-shaped, high acidity tolerant, catalase negative and non-sporulating behavior. Production of lactic acid as a main by-product of glucose fermentation is one of their most well-known attribute; in addition they secrete a number of proteinaceous antimicrobial compounds (bacteriocins) which prevent food from spoilage at the hands of pathogen proliferation (Mokoena, 2017). Health benefits conferred by lactic acid bacteria render them as being regarded as “probiotic” (pro- Latin, for; biotos- Greek, life). These are the microbes (bacteria or yeast) in supplemented or fermented food products which help improve human health. Yogurt and similar fermented foods help supply probiotics to epithelial lining of gut, which is the main site for the growth of intestinal flora (Hati et al., 2013). FAO/WHO designates a microbe as “probiotic” if it is non-pathogenic, confers beneficial effects on host health and is ingested in adequate amount (FAO, 2006).
Lactic acid bacteria are found in a number of habitats. Most important genera of LAB i.e. Bifidobacterium and Lactobacillus are residents of human gastrointestinal tract (GIT). In addition, they are also found in fermented foods and food products (Kumari et al., 2012). Lactic acid bacteria may have two major types of fermentation patterns: (i) the end product can be lactic acid which is a result of complete fermentation of carbohydrates or (ii) it can be a mixture of lactic acid, acetic acid, CO2 and ethyl alcohol (Nuraida, 2015). The former are called homo-fermenters while the latter are known as hetero-fermenters (Nuraida, 2015). LAB isolates from fermented foods have been reported to show more promising probiotic activity than those of the other sources (Rhee et al., 2011). They are majorly found in fermented products, dairy items and GIT. The research regarding the LAB-plant interaction is relatively scarce, although recently, presence of lactobacilli in fruit pulps and processing byproducts has been documented (Garcia et al., 2016). Some other plant sources evaluated for the presence of lactobacilli include fermented olives (Bautista-Gallego et al., 2013), durum wheat (Minervini et al., 2015), rice and paddy rice silage (Ennahar et al., 2003; Ikeda et al., 2013), fresh vegetables like cabbage and cauliflower (Amin et al., 2009) and as phyllosperic entities (McGarvey et al., 2019). The plant sources have not been probed adequately for the isolation of LAB and very few studies have been carried out on the application of plant-associated LAB for growth promotion and disease management in tomato and wheat (Hamed et al., 2011; Suproniene et al., 2015. More prominent microbial group, generally used for such studies, is rhizobacteria (e.g. Rhizobium) which are the major colonizers of plant roots and rhizosphere. These plant growth-promoting rhizobia (PGPR) play key role in agriculture (Majeed et al., 2015).
Chickpea (Cicer arietinum L.) is South Asia’s first and world’s third most important pulse crop. In Pakistan and other developing countries, it is considered as an important legume crop because it is a rich protein source, (Chibbar et al., 2010). There are two types of chickpea: one with usually large sized ram’s-head shaped seed with smooth surface and beige colored coat – the kabuli type (Fig. 1A) whereas the other type – the desi type (Fig. 1B), has smaller, rough surfaced and darker seeds (Pande et al., 2005). Pakistan is the third largest chickpea producer after India (68.7%) and Australia (5.1%), contributing 4.1% of production worldwide (Ferede et al., 2018). During the past few years, chickpea yield has been declined and this decrease in yield is associated with biotic (wilt and blight etc.) and abiotic (drought, salinity) stresses. Blight and wilt of chickpea are of considerable importance. Chickpea wilt (Fusarium wilt) is caused by a soil-borne fungal pathogen – Fusarium. It germinates in response to plant root exudates, penetrates the vascular bundles of chickpea root and blocks the transport of water and minerals to aerial parts of the plants due to its physical growth inside the xylem, hence creates a condition of water deficit know as wilt (Kraft et al., 1993). This disease has a worldwide occurrence and is capable of causing massive yield loss (Sharma and Muehlbauer, 2007). The microbial antagonists including PGPR have been utilized to manage wilt disease and enhance plant growth in pulses (Gholve and Kurundkar, 2002; Kumari and Khanna, 2016). However, application of LAB for biocontrol of wilt disease and promote plant growth in chickpea is not yet documented. Therefore, the present study was designed to explore plant-associated endophytic lactic acid bacteria, enhance their plant growth promoting efficacy by induced mutation and unlock their potential as bio-inoculant for improvement in chickpea crop.
Materials and methods
This research work was carried out in the laboratories of Marker Assisted Breeding (MAB) Group, Plant Breeding and Genetics Division (PBGD) and net house facility located in Nuclear Institute for Agriculture and Biology (NIAB), Faisalabad, Pakistan.
Isolation and characterization
For isolation of lactic acid bacteria, cotton and rice leaves were cut from live plants, packed in clear zip lock bags and instantly brought into the sterilized environment in the lab. Plant leaves were dipped in 0.1% sodium hypochlorite (NaOCl) solution for 1 min for surface sterilization. Afterwards, leaves were cut to small pieces and dispersed in vials containing sterilized Lactobacillus plantarum specific medium (LPSM) (Bujalance et al., 2006), and placed anaerobically in an incubator for 3 days at 37°C. Inoculum from LPSM broth culture was spread over LPSM agar plates in triplicates and incubated. After growth, well separated morphologically distinct colonies were randomly picked and sub-cultured on de Man, Rogosa and Sharpe (MRS) agar (de Man et al., 1960) plate for selection of single pure colony. The selected pure isolates were named (LPA1-LPA7) and maintained on LB agar slants for further confirmation. The characterization of isolates was carried out by Gram staining, microscopy, physiological and biochemical assays (indole, methyl red, Voges-proskauer, citrate, catalase, urease, oxidase, milk coagulation and sugar fermentation test). The selected isolates were identified using the 16S rRNA sequence analysis. Genomic DNA was isolated and PCR products were sequenced with 27f and 1492r primers.
Screening for anti-fungal potential
LAB cultures were transferred on potato dextrose agar (PDA) plates in the form of 3 cm streaks (one streak/plate) and incubated at 37°C for 3 days. After incubation, plugs from growing edge of 5 days old Fusarium solani mat were cut and placed on the agar plate at a distance of 1 cm from the bacterial streak. Plates were left at 37°C for 7 days. Relative reduction in growth was given by following formula:
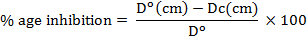
Where, Do is the fungal growth on control plates, and Dc indicates fungal growth on co-cultured plates.
Physical mutagenesis
Fresh culture of a presumptive LAB isolate (LPA6) was prepared by inoculating 10 ml of sterile MRS broth with 0.1 ml of inoculum. Incubation at 37°C was given for 3 days. Afterwards, culture tubes were irradiated by using cobalt-60 (60Co) irradiation source (Gamma Cell 220) available at NIAB for Ɣ-irradiation at various doses (0, 25, 50, 75 and 100 Gy). The process of irradiation was carried out by designated personnel. After irradiation, 30 µl from each tube was spread over MRS agar plates and incubated at 37°C for 3 days. After incubation, survivor colonies were counted and survival percentage was calculated by the formula given in the literature (Tu et al., 2016). The potential survivor mutant colonies were picked and sub-cultured on fresh MRS agar plates for selection of isolated single colony. The selected pure mutant colony was subsequently maintained on agar slant and named as MLPA6.
Preparation of lab inoculum
Fresh cultures of wild type and mutant isolates were prepared in LB broth. Medium was inoculated with microbial culture (0.01% v/v) and incubated at 37°C for 3 days. After incubation, cultures were harvested and spun at 3000 x g for 10 min to get bacterial cells pellets. Inoculum was prepared by suspending cells of both wild and mutant isolates in sterilized 0.1% carboxy-methyl cellulose (CMC) solution. The optical density (OD) of cell suspension was monitored on spectrophotometer (Milton Roy Spectronic 21) at 600 nm.
Net house experiment
Seeds of two chickpea varieties, kabuli-type CM-2008 and desi-type Punjab-2008 (Pb-2008) were selected and treated with LAB cell suspensions (~ 108 CFU/ml) under four treatments; T1 (wild type LPA6), T2 (mutant MLPA-6), T3 (consortium of LPA6 + MLPA6 at 1:1), T4 (untreated seeds). Treated and untreated (control) seeds were sown in micro-plots of net house in December and harvested in April. This experiment was laid out in a Randomized Complete Block Design (RCBD) with three replicates for testing the effect of four treatments on two chickpea varieties (CM-2008 and Pb-2008).
Data analysis
Data was recorded for growth and yield parameters and MS Excel and STATISTIX 8.1 were used for statistical analyses of experimental data.
Results
Isolates and their antifungal activity
Seven presumptive LAB isolates (LPA1 to LPA7) were primarily selected on the basis of their appearance and colony morphology on MRS agar plates (Table I). It was observed that two isolates (LPA1 and LPA7) which
Table I.- Characterization of endophytic lactic acid bacteria isolated from plants.
|
Characteristic |
LAB isolate |
||||||
|
LPA1 |
LPA2 |
LPA3 |
LPA4 |
LPA5 |
LPA6 |
LPA7 |
|
|
Morphology |
|||||||
|
Size |
Small |
Medium |
Medium |
Small |
Small |
Large |
Small |
|
Shape |
Rod |
Cocci |
Rod |
Rod |
Rod |
Rod |
Rod |
|
Margin |
Entire |
Irregular |
Irregular |
Entire |
Entire |
Irregular |
Entire |
|
Color |
White |
Milky |
White |
Milky |
Milky |
White |
Milky |
|
Gram stain |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Biochemical tests |
|||||||
|
Indole |
- |
- |
- |
- |
- |
- |
- |
|
MR |
- |
- |
- |
- |
- |
- |
- |
|
VP |
- |
- |
- |
- |
- |
- |
- |
|
Citrate |
- |
- |
- |
- |
- |
- |
- |
|
Catalase |
- |
- |
- |
- |
- |
- |
- |
|
Urease |
- |
- |
- |
- |
- |
- |
- |
|
Oxidase |
- |
- |
- |
- |
- |
- |
- |
|
MC |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
MR, methyl red; VP, Voges-Proskauer; MC, milk coagulation; +, positive; -, negative .
were obtained from cotton were similar in shape and size (round and small) but different in color. The rest of the five isolates, attained from rice leaves, exhibited variable patterns in colony morphology (Table I). Gram staining and microscopic observation suggested that six out of seven isolates were Gram positive rods whereas only one isolate (LPA2) was Gram positive cocci. Results for biochemical tests were fairly consistent among all isolates. All isolates were negative for indole, methyl-red, Voges-Proskauer, citrate, catalase urease and oxidase tests. However, all seven isolates showed the ability to coagulate milk with acid production. They were also able to ferment different sugars including glucose, fructose and lactose. The bacterial isolates identified using the 16S rRNA sequence analysis were Bacillus tropicus (4 isolates: LPA1, LPA2, LPA5, LPA7) and Bacillus safensis (3 isolates: LPA3, LPA4, LPA6) . All seven isolates were tested for their antagonistic potential against F. solani. Out of seven, four isolates were not considerably active against growing mycelial mat, whereas antifungal activity was observed in three isolates i.e. LPA1, LPA3 and LPA6 (Fig. 2). Among these, LPA6 (Bacillus safensis) was the most effective against F. solani which restricted fungal growth by 25.5% (Table II).
Table II.- Percent (%) inhibition of Fusarium solani by lactic acid bacteria (LAB) on potato dextrose agar (PDA).
|
S. No. |
LAB isolate |
% Inhibition |
|
1 |
LPA1 |
18.9 |
|
2 |
LPA3 |
23.6 |
|
3 |
LPA6 |
25.5 |
Physical mutagenesis
Figure 3 shows the survival percentage of a promising isolate (LPA6) after irradiation with Ɣ-rays. A large number of colonies were observed on control (un-irradiated) plate (Fig. 3A) and culture plates irradiated with low doses i.e. 25 and 50 Gy. The number of colonies drastically dropped to only four with 0.07% survival rate on culture plate irradiated with 75 Gy (Fig. 3B). However, no colonies were observed on 100 Gy plate. Survivor colonies from 75 Gy plate were transferred to fresh LB medium and considered as mutants of LPA6.
Growth and yield parameters
Overall effect of LAB inoculation (T1, T2 and T3) on plant growth and yield in kabuli and desi chickpea varieties (Fig. 4). Variations among four treatments for plant height and root length were observed in both varieties (Fig. 4A, B). In CM-2008 (Fig. 4A), maximum increase in plant
height was observed in MLPA6 mutant (T2) inoculated plants with a value of 16.7%, followed by 11.9% and 9.4% in LPA6 wild type (T1) and consortium of LPA6 and MLPA6 (T3) inoculated plants, respectively as compared
to un-inoculated control (T4) plants. Likewise, maximum increase in root length of kabuli variety was noticed in MLPA6 mutant (T2) inoculated plants. Hence MLPA6 tended to promote plant height as well as root length in kabuli chickpea and manifested an increase of 5.9% in root length over control (Fig. 4A). Similar trend of increase in plant height and root length duet to LAB treatments was demonstrated in Pb-2008 (Fig. 4B). In this desi variety, maximum increase in plant height (16.8%) was attributed to LPA6 wild type (T1) inoculated plants whereas remarkable increase in root length (53%) was manifested in MLPA6 mutant (T2) inoculated plants over control (Fig. 4B). Results indicated the great potential of LAB mutant (T2) for promoting root growth in desi chickpea plants.
None of the bacterial treatments (T1, T2, T3) raised the number of primary branches per plant in CM-2008 (kabuli) as compared to the untreated control (T4) and remained consistent with two branches per plant in each treatment (Fig. 4C). However, the number of secondary branches per plant in kabuli chickpea was considerably increased to 19.1% in MLPA-6 mutant (T2) inoculated plants followed by 13.3% and 8.9% in consortium (T3) and LPA-6 wild type (T1) inoculated plants, respectively over control (T4). In Pb-2008 (desi), the number of primary branches per plant remained the same in all the inoculated and un-inoculated plants (Fig. 4C). Nevertheless, bacterial inoculations tended to improve the number of secondary branches per plant in desi chickpea by 16.9%, 7.9%, 2.3% in LPA6 wild type (T1), MLPA6 mutant (T2) and consortium (T3) treated plants, respectively as compared to control (T4).
A marked increase was observed in pod number and seed number of LAB treated plants as compared to untreated control (T4) in both varieties (Fig. 4D). In CM-2008, increase in pod number per plant over control was 59.5%, 63.2% and 65.1% in LPA6 wild type (T1), MLPA6 mutant (T2) and consortium (T3) treated plants, respectively. Likewise, seed number per plant was also increased in all inoculated plants in comparison to un-inoculated (T4) plants. Maximum increase in seed number was observed in MLPA6 mutant (T2) inoculated plants (75%), followed by consortium (T3) inoculated plants (74.1%) and LPA6 wild type (T1) inoculated plants (68.3%). All LAB inoculations performed equally well in improving the number of pods and number of seeds per plant in Pb-2008 (desi). The percent increase in pod number ranged from 40.2 to 44.7% in inoculated plants (Fig. 4D). The maximum increase was exhibited by LPA6 wild type (T1: 44.7%) inoculated plants which was followed by MLPA6 mutant (T2: 43.5%) and consortium (T3: 40.2%) treated plants. Similar trend of increase in seed number was observed in all LAB treated plants. Maximum percent increase in number of seeds (52.6%) was recorded in LPA6 wild type (T1) inoculated plants whereas MLPA6 mutant (T2) and consortium (T3) treated plants were found to produce 50% and 48.7%, respectively higher seeds than untreated control (T4). Thus LAB inoculations (T1, T2 and T3) produced ~ 50% more seeds in comparison to control (T4) and showed the same pattern of improvement as was noticed in number of pods.
Figure 4E shows that LAB inoculations (T1, T2 and T3) also improved 100 seed weight as compared to control (T4) plants in both varieties. In Kabuli variety, consortium treated (T3) plants produced maximum 100 seed weight (43.14%) whereas increase in 100 seed weight over control (T4) observed in LPA6 wild type (T1) and MLPA6 mutant (T2) plants was 40.9% and 35.5%, respectively. In desi variety (Fig. 4F), MLPA6 mutant (T2) inoculated plants manifested maximum increase (24%) in 100 seed weight whereas LPA6 wild type (T1) produced 13.2% higher than control (T4). Though, pod and seed number was enhanced in consortium (T3) treated plants but in case of 100 seed weight, a very negligible increase (0.2%) over control (T4) was observed. In terms of plant weight in Kabuli variety, all treatments promoted plant growth compared to un-treated control (Fig. 4E); consortium (T3), LPA6 mutant (T2) and LPA6 wild type (T1) inoculations increased plant weight by 90%, 58.3% and 43%, respectively. In desi variety, MLPA6 mutant (T2) treated plants showed maximum increase in plant weight (67.9%), whereas both LPA6 wild type (T1) and consortium (T3) treated plants manifested the same percent increase (51.1%) in plant weight compared to control (T4).
Discussion
This study was designed to investigate the presence of lactic acid bacteria as endophytic components and their potential benefits in improvement of two chickpea varieties (CM-2008 and Pb-2008). Use of LAB specific media led to the isolation of seven presumptive LAB isolates. We observed the presence of LAB as endophytic elements of rice although, the LAB have been previously isolated from rice silage (Ennahar et al., 2003), fermented derivatives of rice (Rhee et al., 2011) and fruits (Azmi and Hashim, 2018). Similarly, presence of LAB in cotton plants is rarely mentioned (McInroy and Kloepper, 1994).
Generally bacteria are present in all habitable environments and they possess several properties through which certain groups can be identified. A group of biochemical tests called IMViC is of pivotal importance in identifying microbial groups on the bases of differential behaviors with regard to these tests. These tests are being in use for characterization of E. coli (Arshad et al., 2006) and for differentiation between E. coli and Enterobacter aerogenes (Hemraj et al., 2013). In the present study, all seven isolates examined were Gram +ve in cell wall structure and manifested negative results for indole, MR, Voger-Proskauer, citrate, catalase, urease and oxidase tests. Our results indicated the characteristics of lactic acid bacteria and were in accordance with a previous study (Islam et al., 2016). Lactic acid bacteria cause coagulation with acid production when incubated in milk (Rahman, 2015); our isolates also fermented milk and thus were considered as presumptive LAB. The rod-shaped isolate (LPA6) employed in this study was identified as Bacillus safensis. It is an aerotolerant organism which can grow in high salt concentrations. It is found to be present in a wide range of habitats ranging from spacecraft assembly facility (SAF; hence its species epithet) (Satomi et al., 2006), which was a point of its original isolation to highly saline deserts to rhizosphere to insect, animal and human gut (Branquinho et al., 2014). Studies have shown this microbe to be capable of fungal biocontrol (Berrada et al., 2012) and probiotic properties (Nath et al., 2012).
Balouiri et al (2016) have discussed number of ways by which anti-fungal activity of microbes can be tested in vitro. One of these methods is known as “cross-streak” method where the candidate microbe is streaked perpendicular to the target microbe (Lertcanawanichakul and Sawangnop, 2011). In this study, a variant of cross-streak method has been used as our target fungal culture was placed on the agar plate near the bacterial streak rather than streaked perpendicular to the LAB streak. Formula for calculating percentage fungal inhibition has been discussed in the literature (Ali-Shtayeh and Abu Ghdeib, 1999). Three isolates (LPA1, LPA3 and LPA6) showed antimicrobial activity and inhibited mycelial growth of a fungal pathogen of chickpea (F. solani) by 23.6% and 25.54%, respectively which is in conformity with previous findings (Husain et al., 2017).
Physical mutagenesis is not widely used for hyper-production of bioactive compounds in lactic acid bacteria. However, few studies have reported the use of UV radiations for physical mutagenesis in E. coli (Arshad et al., 2010) and LAB (Sobrun et al., 2012). Carbon ion beam has also been used for the irradiation of LAB for enhanced lactic acid production (Hu et al., 2017). Previously, gamma radiations have also been used for optimizing prodigiosin compound in Serratia (Elkenawy et al., 2017). In a recent study (Kudryasheva et al., 2017), it was observed that the luminescence of marine bioluminescent bacteria decreases with a gradual increase in temperature and gamma radiation dose. They reported that gamma radiation may affect different bacteria in different ways but one aspect that is fairly common is that their number tends to decrease as the radiation dosage increases.
In the present study, we used four different doses of γ-rays (0, 25, 50, 75, 100 Gy) and obtained a promising LAB mutant isolate (MLPA6) at a dose of 75 Gy with 0.07% survival rate of mutant colonies. After radiation exposure, plates with the least number of LAB colonies were selected and defined as mutant survivors. This denotes to the fact that in a growth medium where a huge number of colonies were present, only a few of them were able to resist gamma radiation stress by making adequate changes to their genetic makeup, hence turning themselves into mutants. In such experiments, low survival percentage (≥ 1%) is preferred for better chance of finding a mutant (Sobrun et al., 2012). Our LAB mutant isolate (MLPA6) was selected at a survival percentage of 0.07% and was applied on chickpea seeds along with its wild type parent for comparing the growth promoting potential of both isolates.
Application of living microbial cultures to the seeds or seed coats before cultivation is known as “biopriming” and this process is usually employed in plant growth promoting rhizobia (PGPR). The biopriming of seeds confers many good outcomes like speedy and uniform germination and escalated crop establishment which leads to better yield and quality (Mahmood et al., 2016). In addition, it helps microbes to adjust in the natural environments. Biopriming can be performed by a variety of different methods e.g. it can be carrier based – where microbial cultures are mixed with a carriers like peat and then applied to the soil (Boonkerd and Singleton, 2002), seed coating – where cultures are coated over the surface of seeds with the use of adhesives (Bardin and Huang, 2003), root dipping – where roots of live plants are dipped into the microbial cultures (Srinivasan et al., 2009), or soil application – where cultures are applied directly to the soil in the root zone (Bashan, 1998). In the present study, seed coating was done by seed dip method and carboxy-methyl cellulose was used as an adhesive. Instances for the use of lactic acid bacteria for biopriming of grains and pulses are scarce. However, the use of LAB as biopriming agents in tomato and their positive effects on tomato growth and as biocontrol agents have been documented (Abdel-Aziz et al., 2014).
A diverse pattern of growth promoting activities of three LAB treatments (T1, T2, T3) comprising LPA6 wild type, MLPA6 mutant and consortium (LPA6 wild type and MLPA6 mutant) was observed in two chickpea varieties. The influence of three different LAB treatments on growth and yield parameters of kabuli variety (CM-2008) clearly indicated that the bacterial inoculum of MLPA6 mutant (T2) exerted maximum impact on plant height, root length, number of secondary branches, pod number and seed number per plant and consequently enhanced these parameters by 16.7%, 5.9%, 19%, 63.2% and 75%, respectively as compared to untreated control (T4). However, the inoculum of consortium (T3) revealed more pronounced effect on 100 seed weight (43.1%) and plant weight (90%) compared to control (T4) which was attributed to the combined effect of both LPA6 wild type and MLPA6 mutant isolates.
During the present study, it was noted that MLPA6 mutant (T2) also performed well in desi variety and induced beneficial effects on plant growth and yield by increasing root length (53%), pod number (43.5%), seed number (50%), 100 seed weight (24%) and plant weight (67.9%) over control (T4). On the other hand, LPA6 wild type (T1) showed more positive effects on plant height (16.8%), number of secondary branches (16.9%) and pod number (44.7%) in desi chickpea. It was interesting to note that all LAB treatments had potentially beneficial liaison with chickpea. Though, wild type and mutant (LPA-6) isolates singly or in combination (consortium of wild type and mutant) promoted plant growth and enhanced grain yield in both chickpea varieties. However, it was obvious from the findings that mutant isolate performed better and greatly influenced the growth.
Our data showed no significant change in number of primary branches under the influence of LAB inoculations. Previously, change in branch number after PGPR inoculations has been documented (Tagore et al., 2013). It might be due to the specific effects of PGPR inoculants on primary branches whereas in our study number of primary branches was not influenced by LAB inoculant and remained the same. Nevertheless, positive effects of LAB inoculation were directed towards enhanced vegetative growth in chickpea which was in the form of increased plant height, root length and number of secondary branches, etc. Moreover, LAB inoculants also improved the number of pods and seeds along with 100 seed weight and plant biomass. These findings indicate the better performance of plants after the bacterial inoculation (Elkoca et al., 2007). Hence the number of pods per plant has direct effect on seed yield in chickpea (Gaikwad and Monpara, 2012) therefore, the number of pods, number of seeds and 100-seed weight were used as selection criteria for assessing the effect of LAB inoculants on improving the yield in chickpea. Our results suggested that these yield components could be used as performance indicator for LAB inoculants in chickpea crop.
This study is the first report which has provided an insight into the potential usefulness of environment friendly microbes comprising wild type lactic acid bacteria and their gamma irradiated mutant as bio-inoculant in chickpea crop for improving its growth and yield without applying any fertilizer. Since this work was conducted in net house therefore, further experimental work and field trials need to be conducted for application of LAB as bio-fertilizer and to establish the impact of LAB bio-inoculant on chickpea and other legume crops for eco-friendly sustainable agriculture.
Conclusions
The findings of this study suggested that lactic acid bacteria (LAB) enhanced plant growth and yield in chickpea. Hence, these microbes could be used as alternative to chemical fertilizers for improving production of chickpea in addition to their use as probiotics in functional food, poultry and cattle.
Acknowledgment
Authors would like to thank Nuclear Institute for Agriculture and Biology College (NIAB-C), Faisalabad, Pakistan for funding.
Compliance with ethical standards
This manuscript does not contain human studies or experiments using animals.
References
Abdel-Aziz, S.M., Moustafa, Y.A. and Hamed, H.A., 2014. Lactic acid bacteria in the green biocontrol against some phytopathogenic fungi: Treatment of tomato seeds. J. Basic appl. Scient. Res., 4: 1-9.
Ali-Shtayeh, M. and Abu Ghdeib, S.I., 1999. Antifungal activity of plant extracts against dermatophytes. Mycoses, 42: 665-672. https://doi.org/10.1046/j.1439-0507.1999.00499.x
Amin, M., Jorfi, M., Khosravi, A., Samarbafzadeh, A. and Sheikh, A.F., 2009. Isolation and identification of Lactobacillus casei and Lactobacillus plantarum from plants by pcr and detection of their antibacterial activity. J. biol. Sci., 9: 810-814. https://doi.org/10.3923/jbs.2009.810.814
Arshad, R., Farooq, S., and Ali, S.S., 2006. Manipulation of different media and methods for cost-effective characterization of Escherichia coli strains collected from different habitats. Pak. J. Bot., 38: 779-789.
Arshad, R., Farooq, S. and Ali, S.S. 2010. Effect of mutations induced by N-methyl-N′-nitro-N-nitrosoguanidine on expression of penicillin G acylase and β-lactamase in wild-type Escherichia coli strains. Ann. Microbiol., 60: 645-652.
Azmi, N.S. and Hashim, A.M., 2018. Phenotypic and molecular characterisations of lactic acid bacteria isolated from malaysian fruits. Pertanika J. Trop. Agric. Sci., 41: 1599-1611.
Balouiri, M., Sadiki, M. and Ibnsouda, S.K., 2016. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal., 6: 71-79.
Bardin, S.D. and Huang, H.-C., 2003. Efficacy of stickers for seed treatment with organic matter or microbial agents for the control of damping-off of sugar beet. Pl. Pathol. Bull., 12: 19-26.
Bashan, Y., 1998. Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv., 16: 729-770. https://doi.org/10.1016/S0734-9750(98)00003-2
Bautista-Gallego, J., Arroyo-López, F., Rantsiou, K., Jiménez-Díaz, R., Garrido-Fernández, A. and Cocolin, L., 2013. Screening of lactic acid bacteria isolated from fermented table olives with probiotic potential. Fd. Res. Int., 50: 135-142. https://doi.org/10.1016/j.foodres.2012.10.004
Berrada, I., Benkhemmar, O., Swings, J., Bendaou, N. and Amar, M., 2012. Selection of halophilic bacteria for biological control of tomato gray mould caused by Botrytis cinerea. Phytopathol. Mediterr., 51: 625-630.
Boonkerd, N. and Singleton, P., 2002. Production of rhizobium biofertilizer. Biotechnology of Biofertilizers. Narosa Publishing House, New Delhi, pp. 122-128.
Branquinho, R., Meirinhos-Soares, L., Carriço, J.A., Pintado, M. and Peixe, L.V., 2014. Phylogenetic and clonality analysis of Bacillus pumilus isolates uncovered a highly heterogeneous population of different closely related species and clones. FEMS Microbiol. Ecol., 90: 689-698. https://doi.org/10.1111/1574-6941.12426
Bujalance, C., Jiménez-Valera, M., Moreno, E. and Ruiz-Bravo, A., 2006. A selective differential medium for Lactobacillus plantarum. J. Microbiol. Meth., 66: 572-575. https://doi.org/10.1016/j.mimet.2006.02.005
Chibbar, R.N., Ambigaipalan, P. and Hoover, R., 2010. Molecular diversity in pulse seed starch and complex carbohydrates and its role in human nutrition and health. Cereal Chem., 87: 342-352. https://doi.org/10.1094/CCHEM-87-4-0342
De Man, J., d. Rogosa and Sharpe, M.E., 1960. A medium for the cultivation of lactobacilli. J. appl. Bacteriol., 23: 130-135.
Elkenawy, N.M., Yassin, A.S., Elhifnawy, H.N. and Amin, M.A., 2017. Optimization of prodigiosin production by Serratia marcescens using crude glycerol and enhancing production using gamma radiation. Biotechnol. Rep., 14: 47-53. https://doi.org/10.1016/j.btre.2017.04.001
Elkoca, E., Kantar, F. and Sahin, F., 2007. Influence of nitrogen fixing and phosphorus solubilizing bacteria on the nodulation, plant growth, and yield of chickpea. J. Pl. Nutr., 31: 157-171. https://doi.org/10.1080/01904160701742097
Ennahar, S., Cai, Y. and Fujita, Y., 2003. Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. environ. Microbiol., 69: 444-451. https://doi.org/10.1128/AEM.69.1.444-451.2003
Ferede, S., Fikre, A. and Ahmed, S., 2018. Assessing the comptetitiveness of smallholders chickpea production in the central highlands of Ethiopia. Ethiopian J. Crop Sci., 6: 51-65.
FAO, 2006. Probiotics in food: Health and nutritional properties and guidelines for evaluation. Food and Agriculture Organization, Rome, Italy.
Gaikwad, S. and Monpara, B., 2012. Selection criteria for yield improvement in chickpea (Cicer arietinum L.) breeding. J. Agric. Res. Technol., 37: 379-385.
Garcia, E.F., Luciano, W.A., Xavier, D.E., da Costa, W.C., de Sousa Oliveira, K., Franco, O.L., de Morais Júnior, M.A., Lucena, B.T., Picão, R.C. and Magnani, M., 2016. Identification of lactic acid bacteria in fruit pulp processing byproducts and potential probiotic properties of selected Lactobacillus strains. Front. Microbiol., 7: 1371. https://doi.org/10.3389/fmicb.2016.01371
Gholve, V. and Kurundkar, B., 2002. Biocontrol of pigeonpea wilt with Pseudomonas fluorescens and Trichoderma viride. Indian J. Pulses Res., 15: 174-176.
Hamed, H.A., Moustafa, Y.A. and Abdel-Aziz, S.M., 2011. In vivo efficacy of lactic acid bacteria in biological control against Fusarium oxysporum for protection of tomato plant. Life Sci. J., 8: 462-468.
Hati, S., Mandal, S. and Prajapati, J., 2013. Novel starters for value added fermented dairy products. Curr. Res. Nutr. Fd. Sci. J., 1: 83-91. https://doi.org/10.12944/CRNFSJ.1.1.09
Hemraj, V., Diksha, S. and Avneet, G., 2013. A review on commonly used biochemical test for bacteria. Innov. J. Life Sci., 1: 1-7.
Hu, W., Chen, J., Wu, Q., Li, W., Liu, J., Lu, D. and Wang, S., 2017. The mutagenesis of Lactobacillus thermophilus for enhanced l-(+)-lactic acid accumulation induced by heavy ion irradiation. Brazilian Arch. Biol. Technol., 60: e17160337. https://doi.org/10.1590/1678-4324-2016160337
Husain, A., Hassan, Z., El-Mabrok, A.S.W., Lani, M.N. and Munir, M.B., 2017. In vitro efficacy of lactic acid bacteria with antifungal activity against Fusarium sp. Cid124-cs isolate from chilli seeds. Int. J. scient. technol. Res., 6: 128-132.
Ikeda, D.M., Weinert, E., Chang, K.C., McGinn, J.M., Miller, S.A., Keliihoomalu, C. and DuPonte, M.W., 2013. Natural farming: Lactic acid bacteria. Sustain. Agric., 8: 3-4.
Islam, K.N., Akbar, T., Akther, F. and Islam, N.N., 2016. Characterization and confirmation of Lactobacillus spp. From selective regional yoghurts for probiotic and interference with pathogenic bacterial growth. Asian J. biol. Sci., 9: 1-9. https://doi.org/10.3923/ajbs.2016.1.9
Kraft, J., Haware, M., Jimenez-Diaz, R., Bayaa, B. and Harrabi, M., 1993. Screening techniques and sources of resistance to root rots and wilts in cool season food legumes. Euphytica, 73: 27-39. https://doi.org/10.1007/BF00027179
Kudryasheva, N., Petrova, A., Dementyev, D. and Bondar, A., 2017. Exposure of luminous marine bacteria to low-dose gamma-radiation. J. environ. Radioact., 169: 64-69. https://doi.org/10.1016/j.jenvrad.2017.01.002
Kumari, A., Catanzaro, R. and Marotta, F., 2012. Clinical importance of lactic acid bacteria: A short review. Acta Biomed., 82: 177-180.
Kumari, P. and Khanna, V., 2016. Biodiversity of Pseudomonas and Bacillus possessing both bioantagonistic and plant growth promoting traits in chickpea rhizosphere. Int. J. Sci. Nat., 7: 153-158.
Lertcanawanichakul, M. and Sawangnop, S., 2011. A comparison of two methods used for measuring the antagonistic activity of Bacillus species. Walailak J. Sci. Technol., 5: 161-171.
Mahmood, A., Turgay, O.C., Farooq, M. and Hayat, R., 2016. Seed biopriming with plant growth promoting rhizobacteria: A review. FEMS Microbiol. Ecol., 92: fiw112. https://doi.org/10.1093/femsec/fiw112
Majeed, A., Abbasi, M.K., Hameed, S., Imran, A. and Rahim, N., 2015. Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol., 6: 198. https://doi.org/10.3389/fmicb.2015.00198
McGarvey, J.A., Hnasko, R.M., Stanker, L.H. and Gorski, L.A., 2019. Use of phyllosphere associated lactic acid bacteria as biocontrol agents to reduce bacterial growth on fresh produce. Google Patents. https://doi.org/10.4315/0362-028X.JFP-19-246
McInroy, J. and Kloepper, J., 1994. Novel bacterial taxa inhabiting internal tissues of sweet corn and cotton. In: Improving plant productivity with Rhizosphere bacteria (eds. M.H. Ryder, P.M. Stephens and G.D. Bowen). CSIRO, Melbourne, pp. 190–238.
Minervini, F., Celano, G., Lattanzi, A., Tedone, L., De Mastro, G., Gobbetti, M. and De Angelis, M., 2015. Lactic acid bacteria in durum wheat flour are endophytic components of the plant during its entire life cycle. Appl. environ. Microbiol., 81: 6736-6748. https://doi.org/10.1128/AEM.01852-15
Mokoena, M.P., 2017. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules, 22: 1255. https://doi.org/10.3390/molecules22081255
Nath, A., Ghosh, S., Chowdhury, R. and Bhattacharjee, C., 2012. Can whey-based Bacillus safensis (juche1) become a food supplement?—growth kinetics, probiotic activity, sensitivity to natural and synthetic antibiotics and synergy with prebiotics and natural antioxidants. Proceedings of International Conference on Recent Advances in Science and Engineering, ICRASE 2012, Hyderabad, India, pp. 51-55. https://doi.org/10.1007/s40034-013-0011-z
Nuraida, L., 2015. A review: Health promoting lactic acid bacteria in traditional indonesian fermented foods. Fd. Sci. Hum. Wellness, 4: 47-55. https://doi.org/10.1016/j.fshw.2015.06.001
Pande, S., Siddique, K., Kishore, G., Bayaa, B., Gaur, P., Gowda, C., Bretag, T. and Crouch, J., 2005. Ascochyta blight of chickpea (Cicer arietinum L.): A review of biology, pathogenicity, and disease management. Austral. J. agric. Res., 56: 317-332. https://doi.org/10.1071/AR04143
Rahman, S., 2015. Probiotic properties analysis of isolated lactic acid bacteria from buffalo milk. Arch. clin. Microbiol., 7: 5-10.
Rhee, S.J., Lee, J.-E. and Lee, C.-H., 2011. Importance of lactic acid bacteria in asian fermented foods. In: Microbial cell factories. BioMed Central, pp. S5. https://doi.org/10.1186/1475-2859-10-S1-S5
Satomi, M., La Duc, M.T. and Venkateswaran, K., 2006. Bacillus safensis sp. Nov., isolated from spacecraft and assembly-facility surfaces. Int. J. system. Evolut. Microbiol., 56: 1735-1740. https://doi.org/10.1099/ijs.0.64189-0
Sharma, K.D. and Muehlbauer, F.J., 2007. Fusarium wilt of chickpea: Physiological specialization, genetics of resistance and resistance gene tagging. Euphytica, 157: 1-14. https://doi.org/10.1007/s10681-007-9401-y
Sobrun, Y., Bhaw-Luximon, A., Jhurry, D. and Puchooa, D., 2012. Isolation of lactic acid bacteria from sugar cane juice and production of lactic acid from selected improved strains. Adv. Biosci. Biotechnol., 3: 398. https://doi.org/10.4236/abb.2012.34057
Srinivasan, K., Gilardi, G., Garibaldi, A. and Gullino, M.L., 2009. Bacterial antagonists from used rockwool soilless substrates suppress Fusarium wilt of tomato. J. Pl. Pathol., 91: 147-154.
Suproniene, S., Semaskiene, R., Juodeikiene, G., Mankeviciene, A., Cizeikiene, D., Vidmantiene, D., Basinskiene, L. and Sakalauskas, S., 2015. Seed treatment with lactic acid bacteria against seed-borne pathogens of spring wheat. Biocontr. Sci. Technol., 25: 144-154.
Tagore, G., Namdeo, S., Sharma, S. and Kumar, N., 2013. Effect of rhizobium and phosphate solubilizing bacterial inoculants on symbiotic traits, nodule leghemoglobin, and yield of chickpea genotypes. Int. J. Agron., 2013: 581627. https://doi.org/10.1155/2013/581627
Tu, R., Jin, W., Wang, M., Han, S., Abomohra, A.E.-F. and Wu, W.-M., 2016. Improving of lipid productivity of the biodiesel promising green microalga Chlorella pyrenoidosa via low-energy ion implantation. J. appl. Phycol., 28: 2159-2166. https://doi.org/10.1007/s10811-015-0783-2
To share on other social networks, click on any share button. What are these?









