Studies on the Mitochondrial tRNA Gene Variants in Patients with Coronary Artery Disease and Pathogenicity Assessment by in silico Analysis
Studies on the Mitochondrial tRNA Gene Variants in Patients with Coronary Artery Disease and Pathogenicity Assessment by in silico Analysis
Fayaz Khan1, Aziz Uddin1, Hisham N. Altayb2, Bibi Nazia Murtaza3,
Sajid Ul Ghafoor1, Faisal Imam4, Saima Iftikhar5, Sadia Tabassum1,
Khushi Muhammad1* and Muhammad Shahid Nadeem2
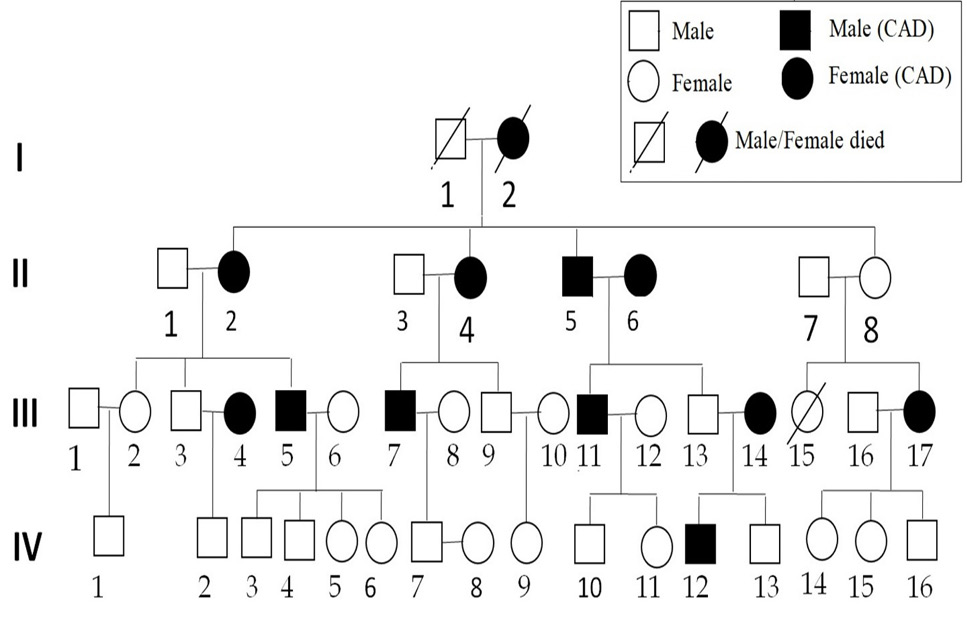
A family pedigree with CAD. Affected individuals are indicated by filled symbols. An arrowhead denotes mutants.
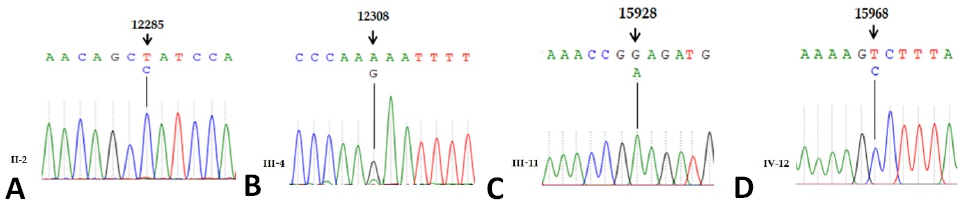
The comparative analysis of affected with revised Cambridge Reference Sequence (rCRS) of human mitochondrial tRNA genes indicating mutations 12285T>C, 12308A>G, 15928G>A and 15968T>C.
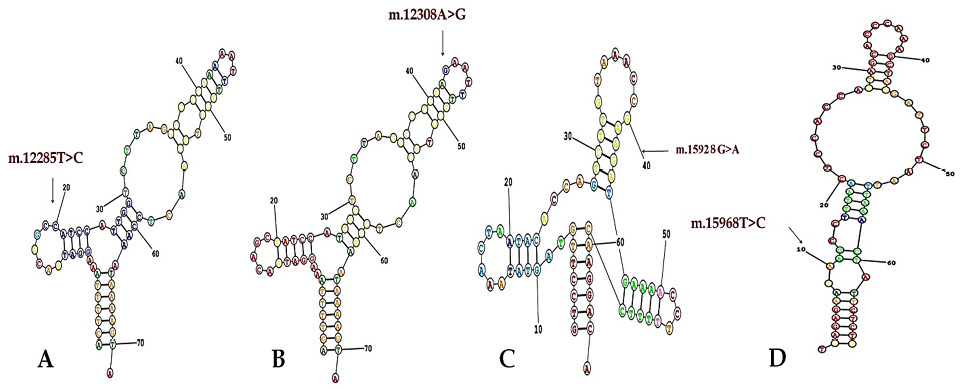
Secondary and tertiary structures of tRNA variants indicating the mutations and 3D structure.
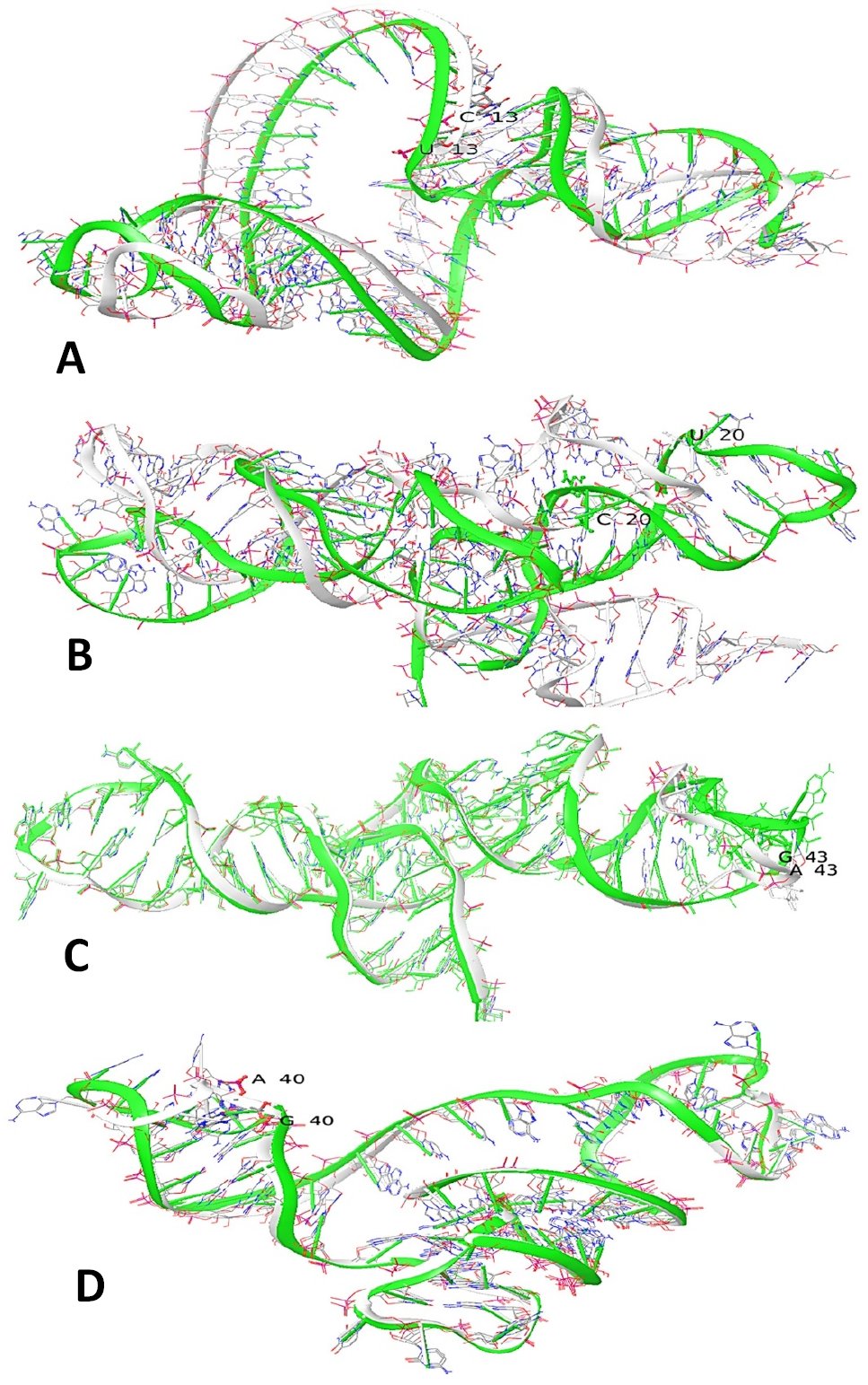
Superimposition of normal (Green) and mutant (White) mtRNA (A). tRNAPro variant U13C (B). tRNALeu2 variant U20C (C). tRNALeu2 variant A43G (D). tRNAThr variant G40A.
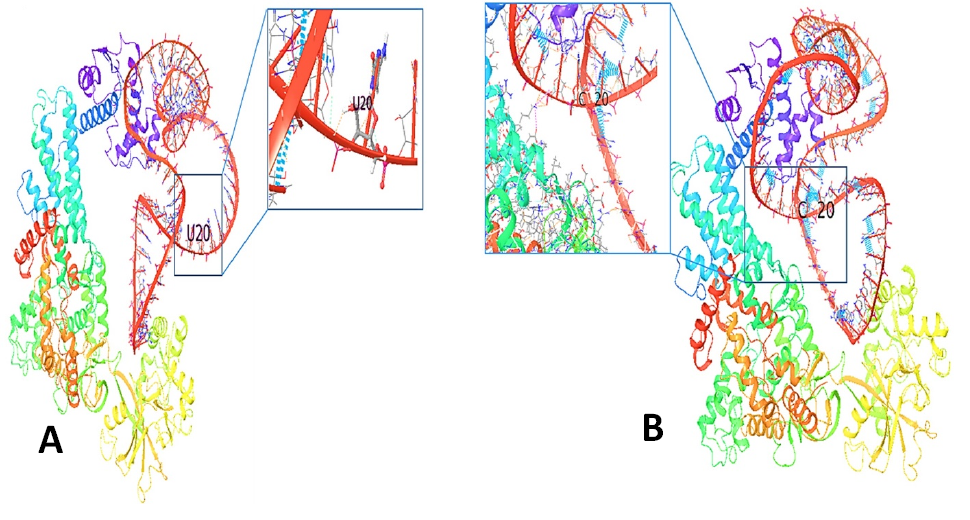
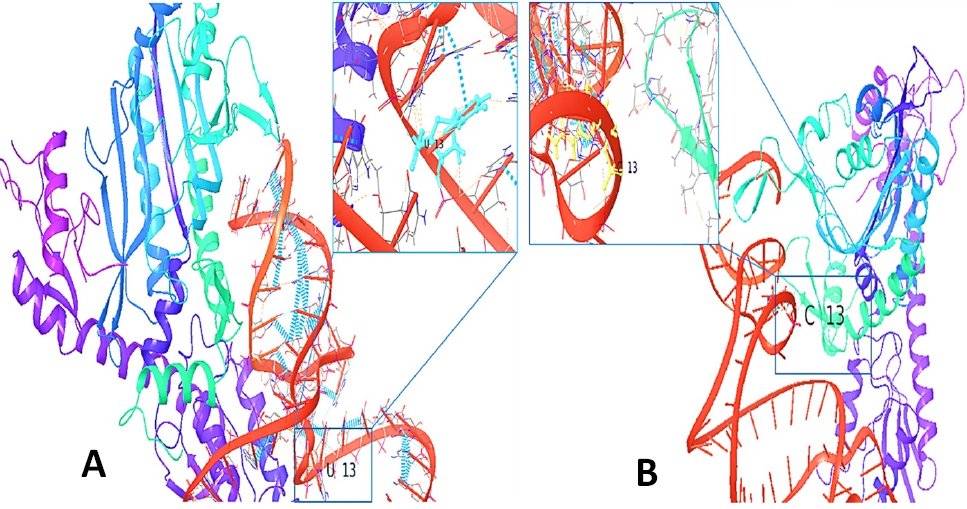
3D structure of the interaction of prolyl- tRNA synthetase with tRNAPro (Red) (A). The normal tRNA Pro with close-up view of the normal residue (U13) (B). The mutant tRNAPro (Red) with close-up view of the mutant residue (C13).
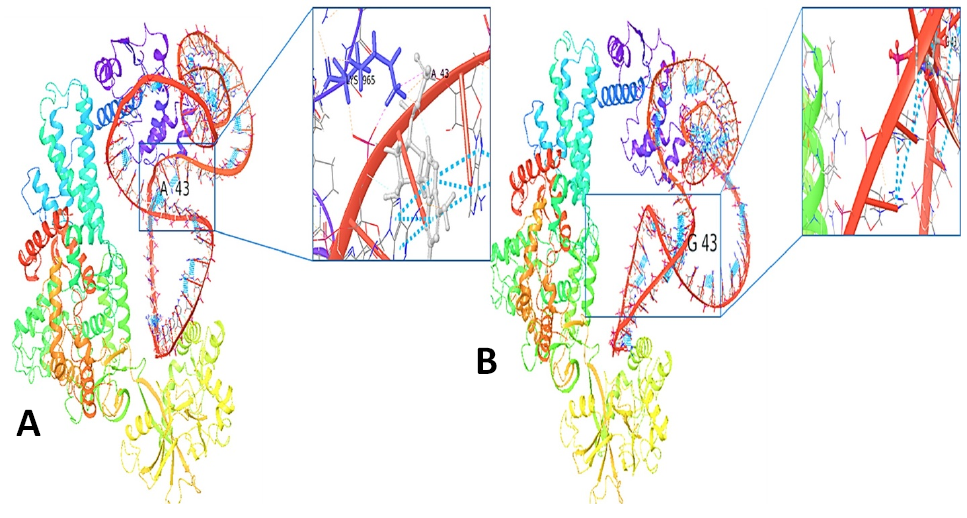
3D structure of the interaction of Leucyl- tRNA synthetase with tRNALeu2 (A) The normal with tRNALeu2 with a close-up view of the normal residue (A43) (B). The mutant with tRNALeu2 with a close-up view of the mutant residue (G43).
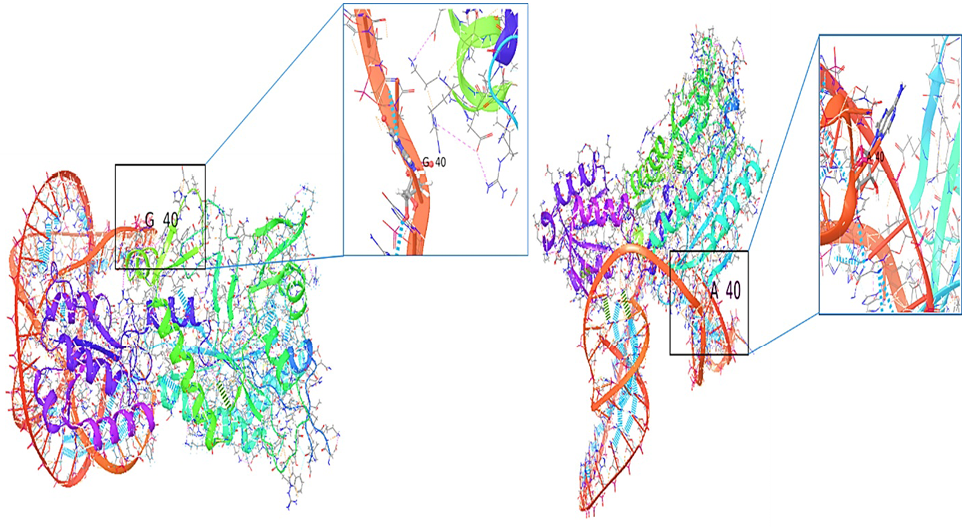
3D structure of the interaction of threonyl- tRNA synthetase with tRNAThr (red) (A). The normal with tRNAThr with a close-up view of the normal residue (G40) (B). The mutant with tRNA tRNA Thr with close-up view of the mutant residue (A40).