A Novel Mutation Ser34Phe in GNRHR causes Hypogonadotropic Hypogonadism during Pubertal Development in Boys
A Novel Mutation Ser34Phe in GNRHR causes Hypogonadotropic Hypogonadism during Pubertal Development in Boys
Misbah Riaz1, Qaiser Mansoor2, Maleeha Akram1, Muhammad Ismail2, Parveen Akhtar3, Shakeel Mirza4, Mazhar Qayyum1, Afzaal Ahmed Naseem1, Faheem Tahir5 and Syed Shakeel Raza Rizvi1*
GNRHR exon 1.1 bands on 2 percent agarose gel. All bands are clear and of proper size of 450 base pairs.
GNRHR 1.1 bands on polyacrylamide gel: Arrow shows the sample with double bands, further sequenced for detection of mutation. All other bands are well defined and single.
Mutated sequence of GNRHR exon 1.1 C-T change at nucleotide position 101 causing Ser34Phe change. Mutation is indicated by arrow.
Normal sequence of GNRHR exon 1.1. Arrow indicates the normal C position.
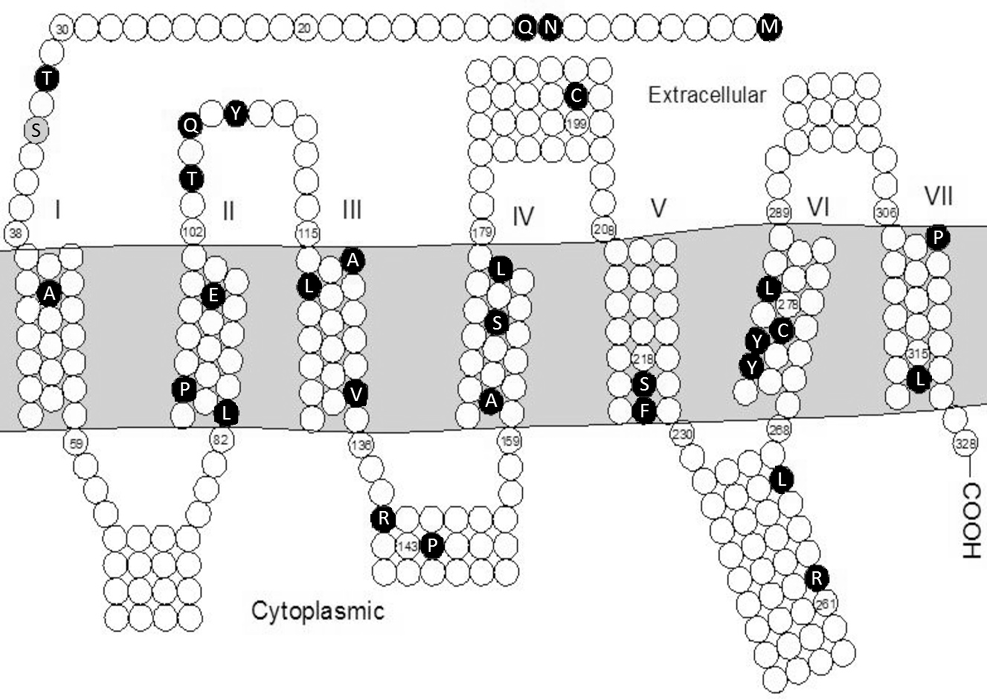
Topology diagram of the amino acid sequence of the human GNRHR protein, showing all mutations depicted so far (black circles with white letters) including Ser34Phe (grey circle with black letter) (deletions are not mentioned).
T-G synonymous mutation in GNRHR exon 1.1 at nucleotide position 123. Arrow indicates the mutated nucleotide. Lines for T (red) and G (black) are superimposed at the mutation.
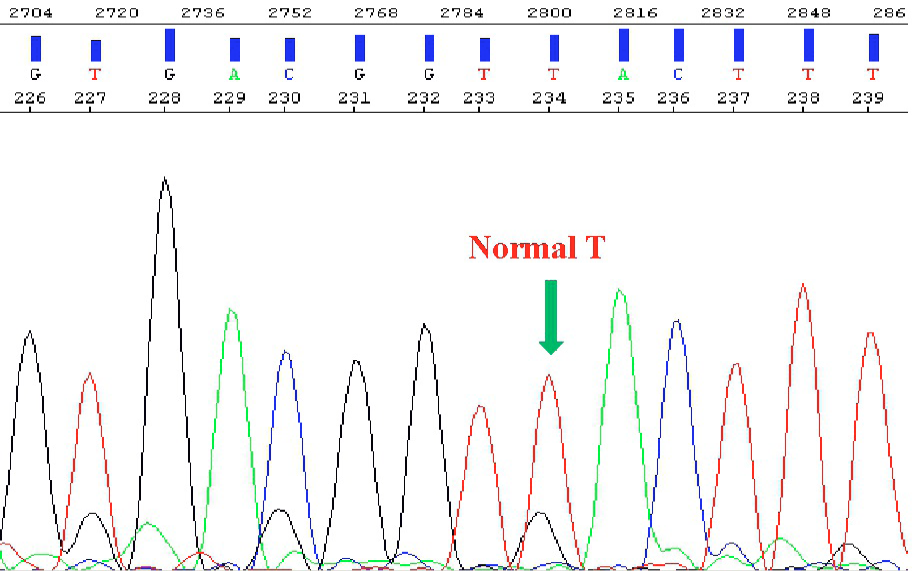
Normal sequence of GNRHR exon 1.1. Arrow indicates the normal T position.