Adaptive Mutations in Nuclear Export Protein and Non-Structural 1 Protein of Avian Influenza a H9N2 Virus Circulating in Punjab, Pakistan
Adaptive Mutations in Nuclear Export Protein and Non-Structural 1 Protein of Avian Influenza a H9N2 Virus Circulating in Punjab, Pakistan
Rehman Shahzad1, Saba Irshad1*, Malik Saddique Mehmood1 and Faisal Amin2
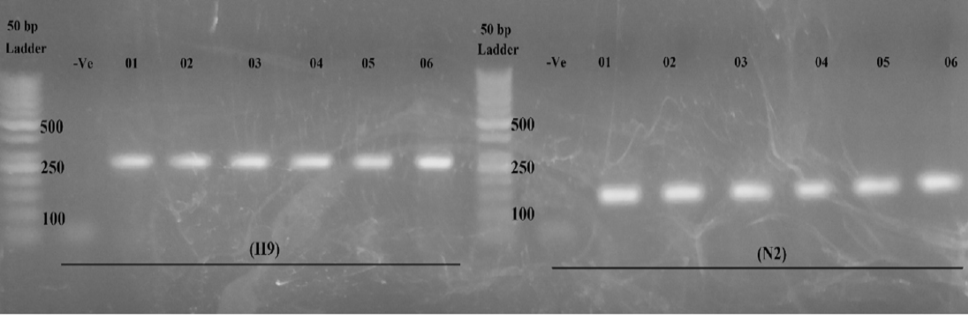
Agarose gel (2%) electrophoresis for six H9N2 RT-PCR positive samples. First lane 50 base pair DNA ladder, second lane is negative control followed by six positive samples of H9N2 virus.
Agarose gel (1%) electrophoresis for amplified and purified segment 8 of six H9N2 virus. First laneis 100 base pair DNA ladder, followed by six lanes with purified segment 8 of 840 base pairs.
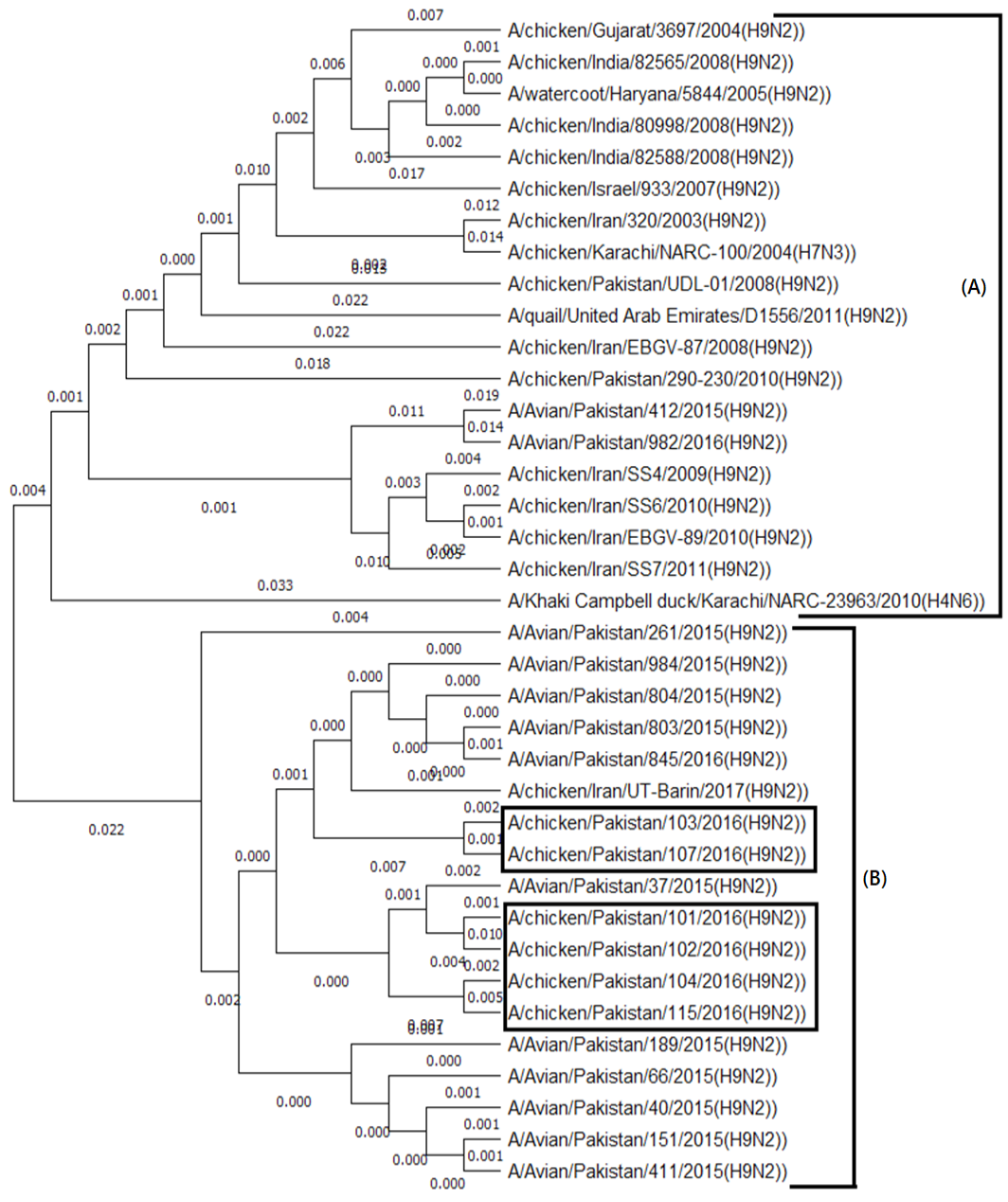
Neighbor-Joining phylogenetic tree constructed using MEGA X (version 10.0.5) software with Tamura-Nei model and 1000 bootstrap values.
Multiple Sequence Alignment (MSA) analysis by online server Clustal Omega: NEP sequnces alligned and compared with the sequence of wild type AVX 27241(grey colour), mutations are highlighted in red colour.
Multiple Sequence Alignment (MSA) analysis by online server Clustal Omega: NSI sequences alligned and compared with wild type AVX 27241 (grey colour) mutations are labelled in red colour.