BmARM-Like Protein from Silkworm, Bombyx mori (Lepidoptera) is Putatively Involved in Response against BmNPV Infection
BmARM-Like Protein from Silkworm, Bombyx mori (Lepidoptera) is Putatively Involved in Response against BmNPV Infection
Xue-yang Wang1, Shang-zhi Zhang1, Ming-hui Liu2, Dong Yu1, Yan Ma1, Dong-qiong Fei1, Hai-zhong Yu1 and Jia-ping Xu1,*
cDNA sequence and deduced amino acid sequence of BmARM-like. The start and termination codons are denoted in the black frame. The functional domains of BmARM-like protein are underlined.
Sequence comparisons of BmARM-like and its homologs. Identical amino acids are highlighted in mazarine, and the positive amino acids are highlighted in pink and wathet. The functional domains of BmARM-like protein are underlined.
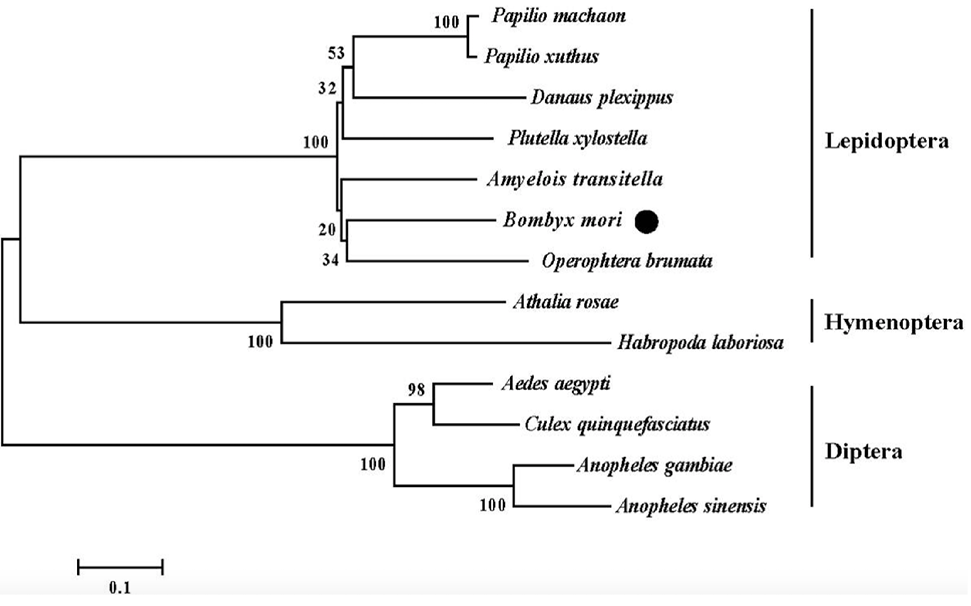
Phylogenetic analysis of the BmARM-like amino acid sequence using the published sequences of ARM-repeat proteins from other species. The tree was constructed using the neighbor-joining method with pairwise deletion of gaps in MEGA 5. Numbers on nodes indicate bootstrap values from 1000 replicates.
RT-qPCR analysis of the expression patterns of BmARM-like mRNA. A, B, C, D, and E show the expression patterns of BmARM-like in different silkworm tissues (A), different developmental stages (B), during egg development (C), during pupal development (D), and during molting (E). F and G show the expression patterns of BmARM-like in the midgut (F) and hemolymph (G) of different resistant silkworm strains following BmNPV infection. Data was normalized using BmGAPDH and is represented as mean ± standard error of the mean from three independent experiments. Different letters on the bars indicate statistically significant differences based on ANOVA followed by Tukey’s HSD multiple comparison test (p < 0.05).
Western blot analysis of the specificity of polyclonal antibodies to BmARM-like protein. Samples were separated by 12% SDS-PAGE under reducing conditions. M marker; 1,2,3 SDS-PAGE; 1’,2’,3’ Western blotting; lane 1,1’ lysate from E.coli without induction, lane 2,2’ lysate from E.coli following induction with IPTG, lane 3,3’ total protein extracted from silkworm. 71 kDa band in silkworm and 31.1 kDa band in E.coli were correspond to BmARM-like protein.
Immunofluorescence analysis of BmARM-like protein distribution in different silkworm tissues. a–t, negative controls for A–T, respectively. a–d and A–D, head; e–h and E–H, midgut; i–l and I–L, fat body; m–p and M–P, testis; q–t and Q–T, ovary. Trans (white), optical transmission; DAPI (blue), nuclear staining; FITC (green), BmARM-like protein staining; Merge, merged images for DAPI and FITC. Scale bars = 200 μm.
Representative western blot analysis of the expression of BmARM-like protein in the midguts of different silkworm strains following BmNPV infection. β-actin was used as a loading control. M, marker. Data shown are representative of three independent experiments.