Candidate Chemosensory Genes Identified in the Procambarus clarkii (Decapoda: Cambaridae) by Antennules Transcriptome Analysis
Candidate Chemosensory Genes Identified in the Procambarus clarkii (Decapoda: Cambaridae) by Antennules Transcriptome Analysis
Zhengfei Wang1*, Chenchen Shen1,2, Yiping Zhang1, Dan Tang1,3, Yaqi Luo1, Yaotong Zhai1, Yayun Guan1, Yue Wang1 and Xinyu Wang1
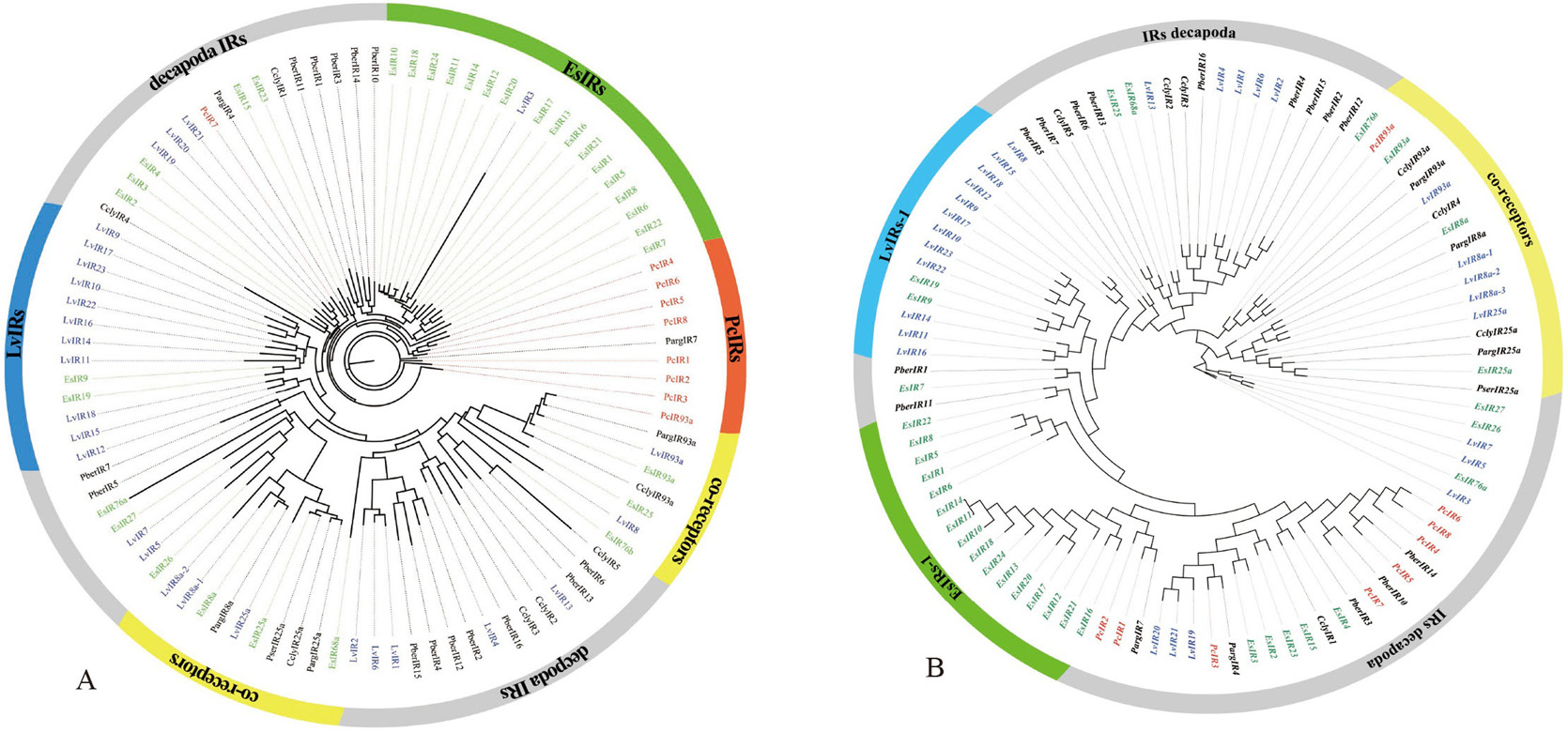
Phylogenetic trees of PcIR sequences with other Decapoda IR sequences, A is the ML phylogenetic tree, and B is the NJ phylogenetic tree. Procambarus clarkii (red), Litopenaeus vannamei (blue) and Eriocheir japonica sinensis (green), and other Decapoda IRs font are colored black. Moreover, out ring containing IR co-receptors are colored yellow.
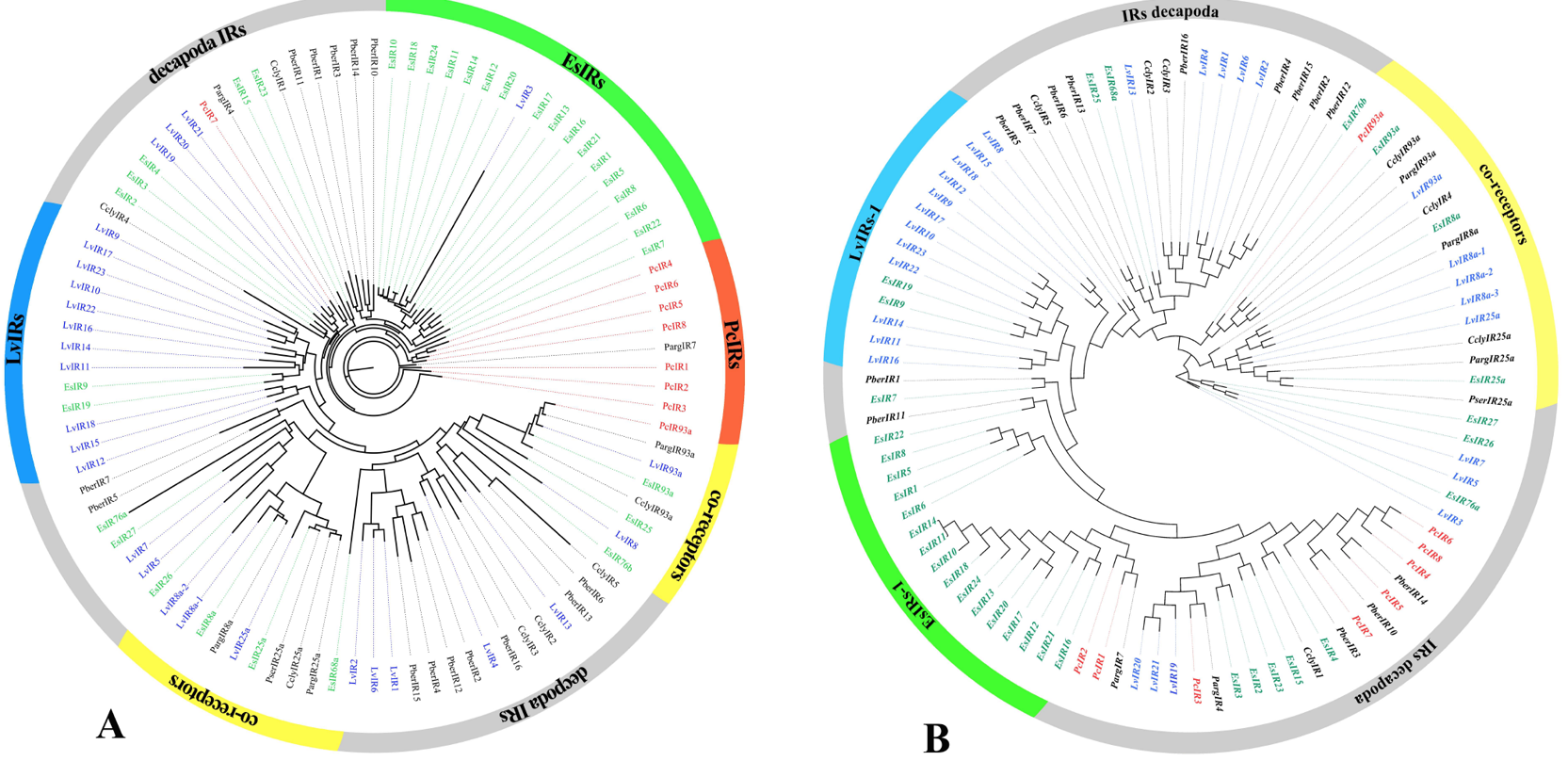
Phylogenetic trees of candidate ionotropic receptors (IRs, IGluRs, and VIGluRs), A is the ML phylogenetic tree and B is the NJ phylogenetic tree. Includes sequences from Procambarus clarkii, Litopenaeus vannamei, Eriocheir japonica sinensis, and other Decapoda. IRs font is colored red; IGluRs font is colored blue; VIGluRs font is colored black. Moreover, the region containing IR co-receptors are colored yellow or orange.
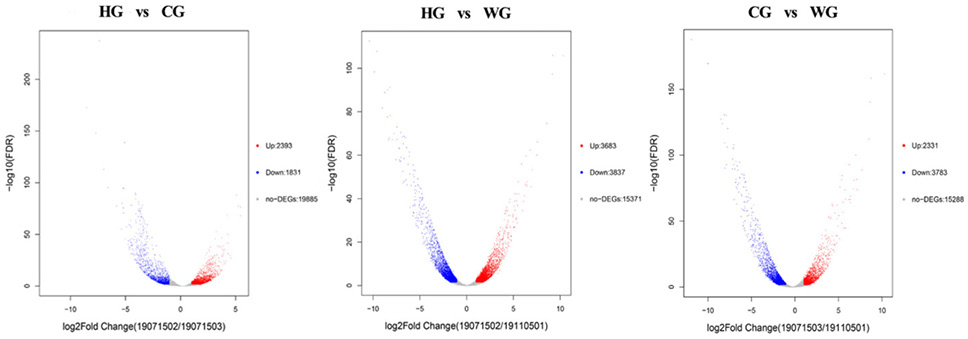
Volcano plot showing the gene expression differences between the WG group, HG group, and CG group. Each dot in the graph represents a specific gene or transcript. Red dots represent genes that are significantly up-regulated, blue dots represent genes that are significantly down-regulated, and gray dots represent genes that are not significantly different points on the left and top represent more significant expression differences.
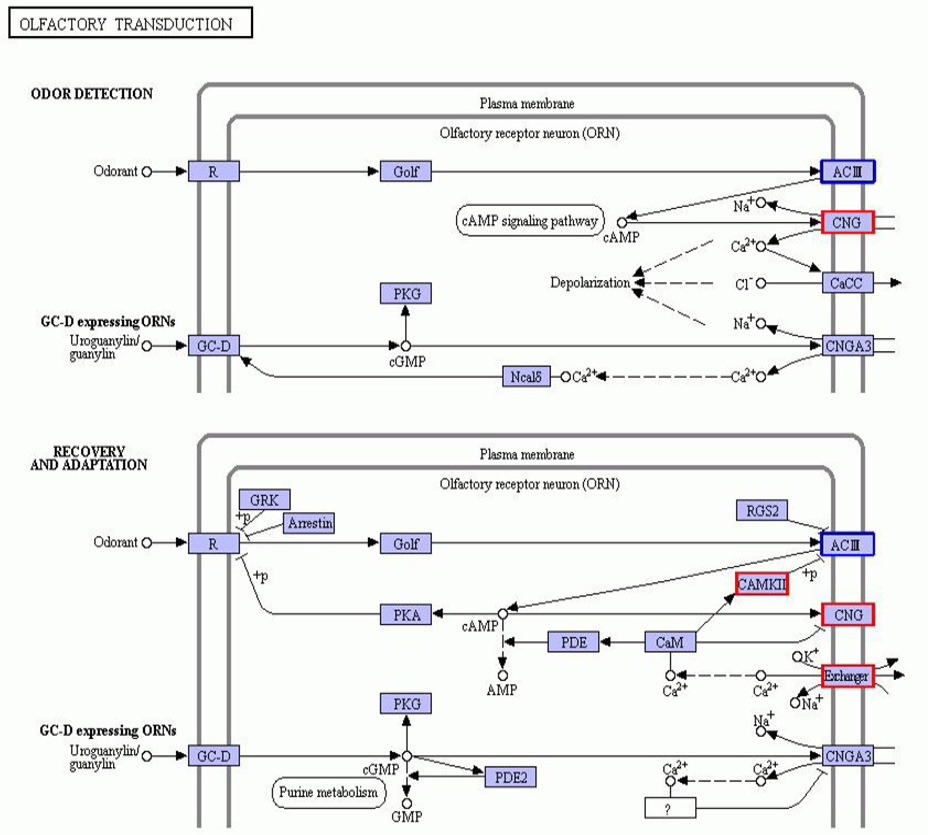
Annotation diagram of up and down regulated gene KEGG pathway in the HG and CG group. The genes with red/blue borders in the figure belong to the differential genes detected by this sequencing, where red represents up-regulated genes and blue represents down-regulated genes.