Comparison of Genetic Diversity between the Ex-Situ Conservation Herd and Smallholders of Turkish Grey Cattle
Comparison of Genetic Diversity between the Ex-Situ Conservation Herd and Smallholders of Turkish Grey Cattle
Süleyman Kök*
Department of Genetics and Bioengineering, Faculty of Engineering, Trakya University, TR-22030, Edirne, Turkey
ABSTRACT
Turkish grey cattle (TGC) are facing the danger of extinction. Intensive breeding program has mainly been used for purebred TGC that are under the conservation of Bandırma Livestock Research Institute (BLRE) ex-situ program. This study aims to compare 3 SNP of the genetic features of purebred TGC that are under the conservation of ex-situ (51 cattle) program with the ones raised by the smallholders (79 cattle) in the villages. According to the estimated average heterozygosity values for ex-situ breeding program and smallholders in the villages, the difference between the cattle were found meaningful (P<0.05). The observed average heterozygosity (Ho) value was calculated as 0.4077±0.1922, while the expected heterozygosity (He) value was found 0.3909±0.1663. The research findings show that the difference between the two TGC groups in terms of Calpastatin gene (CAST) loci (P<0.01) and Calpain gene (CAPN1) loci (P<0.05), gene assortment was found to be meaningful and these are incompatible along with Hardy-Weinberg theory. In addition, significance check was done for the expected heterozygosity results for the two TGC groups (BLRE ex-situ and the smallholders in the villages). For the Fis value, the difference was significant for CAPN1 316 loci (P<0.001) and CAST loci (P<0.05) and non-balanced but the CAPN1 4751 loci (P>0.05) which did not display a significant difference. The Fis inbreeding coefficients being negative in the sample populations for CAST loci imply heterogeneity in CAST loci (P<0.001). The effective allele number and allele density are other criteria to show the spatial heterogeneity in a population. The average effective allele number (ne) per loci for TGC samples from the smallholders in the villages, TGC ex-situ conservation herd and the total population sample were calculated as 1.7379±0.3559, 1.6026±0.4422 and 1.7103±0.4018, respectively.
Article Information
Received 30 January 2017
Revised 27 February 2017
Accepted 31 March 2017
Available online 24 July 2017
Key words
Grey cattle, Heterozygosity, PCR, CAPN1, CAST, Ex-situ conservation.
DOI: http://dx.doi.org/10.17582/journal.pjz/2017.49.4.1421.1427
* Corresponding author: [email protected]
0030-9923/2017/0004-1421 $ 9.00/0
Copyright 2017 Zoological Society of Pakistan
INTRODUCTION
In Turkey where there is a much smaller range of farming environments divided mainly into smaller farms, beef is produced primarily as a by-product of milk production and the cattle are mainly dual purpose for milk and beef (Bozkurt, 2012).
According to the recent zoological research, the origin of Turkish grey cattle (TGC) is believed to be a subpopulation of Bos taurus primigenius (Sasimowski, 1987). It is Ukrainian cattle of steppe origin and it is believed to be a relative of some of the local grey cattle living in Europe (Hristov et al., 2014; Soysal and Kök, 2006). They are also quite resistant to pests and illness besides unfavorable climate and nature conditions. They have the ability to survive, feed and breed without human intervention. They spend the whole year, including winter, as free herds. TGC are endangered because of the uncontrolled cross breeding exercise and low productivity (Soysal et al., 2005). A herd of TGC are under the conservation of Bandirma Livestock Research (BLRE) ex-situ program. In additional to a herd of TGC is protected in BLRE is preserved as an in-situ conservation including 12 herds in the 5 provinces by Republic of Turkey Ministry of Food, Agriculture and Livestock.
Heterozygosity indices are important in order to determine genetic differentiation. The impacts of genetic groupings on a population can be determined by looking at heterozygosity levels without needing allele frequencies among different kinds of organisms.
In ınvestigation of genetic structures of the local cattle breed in Turkey, Özkan (2005) obtained quite high Ho values for East Anatolian Red cattle (0.6653), TGC (0.6823) and Anatolian Black (0.7347). Altınalan (2005) found heterozygosity values as 0.433 and 0.449 for TGC and East Anatolian Red cattle, respectively. Özşensoy and Kurar (2014) researched that the observed (Ho) and expected (He) heterozygosity values of the indigenous cattle breeds in Turkey, as a result of they reported that the highest averages range from 0.619 to 0.852, and 0.669 to 0.877, respectively. In their study of miDNA, Kurar et al. (2011) determined the average He values for Anatolian Black, Turkish Grey, South Anatolian Red, East Anatolian Red and Zavot cattle as 0.761, 0.686, 0.758, 0.768 and 0.747, respectively and stated that these values are quite high for such culture races. They also claimed that the cause of the high heterozygosity values can be that the local cattle breeds live closer to the evolution area compared to European cattle breeds and that the local cattle breeds in Turkey have not been exposed to any selection process (Kurar et al., 2011). Savaşçı and Atasoy (2016) specified the Ho index values that were observed in CAST gene for TGC (0.4423) and East Anatolian Red cattle (0.4706) at similar levels with a more heterogeneous structure compared to the local Black cattle (0.500). They also reported quite high values for East Anatolian Red (0.4412) and Black cattle (0.4306) as the best estimated and sampling-error free average Ho value of the genetic variation in the populations while the TGC (0.2788) were found to have lower values compared to other populations, which shows that they might be closely related and more inbred compared to other populations.
Gene migration is the transfer of animals between subpopulations whereas gene flow is the inclusion of new genes in a certain subpopulation. However, if closed breeding is applied to a population, homozygosity can increase as a result of inbreeding.
The fixation index (Fis) is defined as the coefficient of average consanguinity observed in subpopulations and describes the correlation between homologous alleles in the consanguineous individuals within a subpopulation. In other words, it is the probability of two gametes combined by chance both sharing a common ancestor in the groups.
In my study, the genetic variations between the herds of TGC under ex-situ conservation and the smallholders in the villages were compared with respect to the effective allele number (ne), Ho and He values of TGC groups.
Materials and Methods
Animal samples and genotyping
Tissue samples were taken from 79 pure TGC that were raised in the villages of Çanakkale and Edirne Provinces during 2013 and then the samples were taken to Keşan Slaughter House to be used as research material in addition to the blood samples of 51 pure TGC within the ex-situ protection program from BLRE.
DNA amplification and genotyping
Genomic DNA of the blood and tissue samples taken from 130 purebred TGC was isolated using Fujifilm Quick Gene Mini80 device and commercial kits. Spectrometric A260/280 method was used to determine the amount of the DNA that was isolated. In order to determine CAST gene SNP (GenBank Accession No. AY_008267.1:g.282C>G), Polymerase Chain Reaction-Restriksiyon Fragment Length Polymorphism (PCR-RFLP) method was used based on the work of Schenkel et al. (2006). Identification of Calpain gene CAPN1 316 SNP (GenBank Accession No. AF_252504.2:g.5709C>G) with PCR-RFLP method and CAPN1 4751 SNP (GenBank Accession No. AF_248054.2:g.6545C>T) genotype was identified by Amplification Refractory Mutation System-Polymerase Chain Reaction (ARMS-PCR) method was used based on the work of Rinco’n and Medrano (2006). The primers (Sentegen Biotech, Ankara / Turkey) used and fragment sizes that were reproduced (Table I) by Bioneer My Genie 96 Thermal Block PCR device (Bioneer Corporation, South Korea).
For identification of CAST SNP and CAPN 316 SNP genotypes were worked with PCR-RFLP method. CAST PCR products were amplified 523 bp fragments. The PCR products of CAST gene were cut with RsaI (3 U) restriction endonuclease enzyme (REE) (New England Biolabs).
Table I.- The PCR method used, primer sequences and amplification products.
| Marker | Sequences of the primers (5' - 3' ) | bp*** |
|
*CAST |
1Fop:CTCGACTGCGTACCAATTCCGAAGTAAAGC CAAAGGAACA |
523 |
|
2Rop: ATTTCTCTGATGGTGGCTGCTCACT |
||
|
*CAPN1 316 |
1Fop: GCTGTGCCCACCTACCAGCATC |
446 |
|
2Rop: CAGGTTGCAGATCTCCAGGCGG |
||
|
**CAPN1 4751 |
1Fop: CCTGGAGTCCTGCCGCAGCATGGTCAAC |
334 |
|
2Rop: AAGCTGCAGGAGCTGCCCAAAGCCAGGC |
||
|
3Fip: GCATCCTCCCCTTGACTGGGGGGAAACCC |
158 | |
|
4Rip: GTCACTTGACACAGCCCTGCGCCGCA |
231 |
*, RFLP; **, ARMS method; ***, PCR product size (bp); 1Fop, forward outer primer; 2Rop, reverse outer primer; 3Fip, forward inner primer; 4Rip, reverse inner primer.
CAPN 316 SNP fragment of 446 bp amplified using the PCR protocol was incubated with BtgI (3 U) REE (New England Biolabs) for 4 h at 37 oC. CAPN 316 SNP fragment sizes and genotypes obtained are shown in Figure 1.
All PCR reactions were performed EmeraldAmp GT PCR master mix (Takara, Japan). Amplification of DNA was run in the 3 % horizontal gel electrophoresis by “Thermo Scientific electrophoresis and power supply” and then was used for the genotyping by “DNR BioImaging Systems Minibis Pro. Jerusalem, Israel” and software was used for molecular analysis (Image Aide from Spectronics Corporation). The characteristics of TGC animal samples and genotyping methods have been used by Kök et al. (2017).
Statistical analyses
Two subgroups were formed, one for the cattle raised in villages and the other one for the in ex-situ conservation herds within the total TGC sample. The groups were examined with regard to the population genetics considering the environmental interaction. For each loci, the Observed (Ho) and the Expected (He) Heterozygosity in the total sample population and the two TGC groups (Bandırma ex-situ and the smallholders in the villages) were calculated with Levene (1949) unbiasedly according to Nei (1987) and the Fixation Index (Fis) values were calculated according to Wright (1965) and the effective allele number (ne) values were calculated according to Kimura and Crow (1978) (Tables II, III, VI).
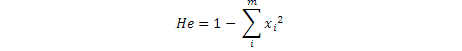
Where, xi is ith allele freqency, m is Number of alleles, He is the expected heterozygosity ratio in a sample population at Hardy Weinberg equilibrium and Ho is number of observed heterozygotes / number of total sample.
ne = 1/Σxi2
Where, ne is Number of effective alleles per loci and xi is the average allele density at the ith loci.
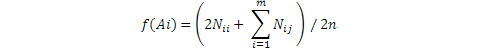
Where, f(Ai) is the ith allele density, n is Number of individuals in the population, Nii and Nij are the number of Aii and Aij genotypes in the population and m is to represents the number of alleles
Fis = (Hs – Hi) /Hs = 1- (Hi/Hs)
Hi = ΣHoj / s
Hs = ΣHej / s
Where, Hoj is the rate of heterozygosity observed in jth population, Hej is the expected heterozygosity of the population j and s is for population number.
The significance control for the difference between the estimated average heterozygosity indices for the two TGC groups was held with respect to probability a in t test table. χ2 test (Degree of Freedom = Allele Number - 1) was used for the significant control of the inbreeding coefficient (Fis) was estimated in each loci and whether the populations show Hardy-Weinberg (HW) equilibrium was discussed. The evaluations were made using Popgene32 version 1.31 program (Yeh et al., 2000).
Results and Discussion
Breeding by dividing populations into local groups helps remove the probability of aleatory copulations and reproduction within a herd. For that reason, homozygosity is more likely to be present in the next generation. BLRE ex-situ conservation herd is a closed herd made up of purebred TGC collected from different villages of the various cities in Marmara Region. This herd is expected to be homozygous. Therefore, this herd was regarded as a subgroup and compared to the Grey cattles in the smallholders in the villages.
In order to determine the effect of environmental interaction on BLRE ex-situ conservation herd and the samples taken from smallholders, genotypic results of the two sample groups were statistically evaluated. In both of the sample the herds of TGC under ex-situ conservation and the smallholders in the villages, in order to eliminate the errors in He value, which is the statistical measurement of genetic variation in all three loci, according to Nei (1987) was estimated to be unbiased.
The Fis value varies between 0 and 1 and if it is negative it shows excess for heterozygosity. If it is close to zero, it implies the presence of Hardy-Weinberg equilibrium and if it is positive, it means excessive homozygosity. Homozygosity index (fixation index) is also defined as the positive deviation of the expected heterozygosity in Hardy-Weinberg rates in a population where inbreeding is applied. Negative Fis value is also defined as the heterogeneity rate which is occurring to the result of remote inbreeding in a certain loci (Yeh et al., 2000). Fis value gives us the percentage of which the heterozygous individuals fall below or above the normal amount in a population. The Fis inbreeding coefficient results according to chi-square (χ2) test values presented in Tables II, III and IV are discussed below. With respect to the inbreeding coefficients computed in the three polymorphic loci in Grey cattle, the differences in CAST (P<0.001) loci were found significant and was not balanced for all population and subgroups (Tables II, III, IV). The Fis inbreeding coefficients being negative in the sample populations for CAST loci imply heterogeneity in CAST loci. While the differences for CAPN1 4751 loci in both the total population and the subgrous (the herds of TGC under ex-situ conservation and the smallholders in the villages) were found nonsignificant (P>0.05) and balanced, the difference for CAPN1 316 loci in BLRE ex-situ population was found significant (P<0.05) and non-balanced. The inbreeding coefficients calculated for other populations were not statistically meaningful. According to Savaşçı and Atasoy (2016) the Fıs value, which was also the indicator of the mean heterozygosity excess determined for TGC, was determined as 5.7% (-0.057) and the population was not-balanced. According to Özşensoy et al. (2010) and Altınalan (2005) for TGC the Fıs value 0.05524 and 0.11930, respectively, these values are insignificant and the populations are balanced.
Table II.- Results for the effective allele number (ne), the observed (Ho) and the expected (He) heterozygosity indices and the fixation index (Fis) tested in CAST and CAPN1 loci in 79 TGC samples in the smallholders in the villages.
| The loci |
ne |
1Ho |
1He |
2He |
3Fis |
| CAST |
1.9920 |
0.6329 |
0.5012 |
0.4980 |
-0.2709*** |
| CAPN1 4751 |
1.8906 |
0.4557 |
0.4741 |
0.4711 |
0.0327ns |
| CAPN1 316 |
1.3311 |
0.2405 |
0.2503 |
0.2488 |
0.0332ns |
| Average |
1.7379 |
0.4430 |
0.4085 |
0.4059 |
|
| ±St. Deviation |
0.3559 |
0.1965 |
0.1377 |
0.1368 |
1, Levene (1949); 2, Nei (1987); 3, Wright (1965); ***, (P<0.001) significance; ns, non- significant.
According to HW equilibrium, while there was upward trend of heterozygous cattle (hyper-heterozygosity ) at 27.09 % in CAST loci (P<0.001), but there were downward trend of heterozygous cattle (hypo-heterozygosity) at 3.27 % and 3.32 % in CAPN1 4751 and CAPN1 316 loci respectively and these heterozygosity differences were found nonsignificant (P> 0.05) in TGC in the smallholders (Table II).
Table III.- Results for the effective allele number (ne), the observed (Ho) and the expected (He) heterozygosity indices and the fixation index (Fis) tested in CAST and CAPN1 loci in 51 TGC samples in BLRE ex-situ conservation herd.
| The loci |
ne |
1Ho |
1He |
2He |
3Fis |
| CAST |
1.6862 |
0.4510 |
0.4110 |
0.4070 |
- 0.1082** |
| CAPN1 4751 |
1.9969 |
0.4902 |
0.5042 |
0.4992 |
0.0181ns |
| CAPN1 316 |
1.1245 |
0.1176 |
0.1118 |
0.1107 |
- 0.0625* |
| Average |
1.6026 |
0.3529 |
0.3423 |
0.3390 |
|
| ±St. Deviation |
0.4422 |
0.2047 |
0.2050 |
0.2030 |
1, Levene (1949); 2, Nei (1987); 3, Wright (1965); **, (P<0.01); *, (P<0.05) significance; ns, non-significant.
In the samples TGC in BLRE ex-situ group, according to HW equilibrium, while there were hyper-heterozygosity at 10.82 % and 6.25 % in CAST and CAPN1 316 loci, respectively, there was hypo-heterozygosity at 1.81 % CAPN1 4751. The expected deviations in the HW equilibrium in CAST loci (P<0.01) and CAPN1 316 loci (P<0.05) in BLRE ex-situ conservation herd was found significant (Table III).
In the total TGC sample, the percentage of hyper-heterozygosity was 17.37 % in terms of CAST loci and this difference was significant (P<0.001). The observed hypo-heterozygosity rates were found 4.52 % and 2.97 % in CAPN1 4751 and CAPN1 316 loci, respectively and the difference was found nonsignificant (P>0.05). Therefore, the expected homozygosity in the total TGC sample population for CAST, CAPN1 4751 and CAPN1 316 loci are 51.97 %, 50.67 % and 80.10 %, respectively. The most homogenous genes in the cattle were found in CAPN1 316 loci (Table IV).
Without the effect of sampling error, the average He values in order to estimate the genetic variation in the populations were calculated as 0.4059±0.1368 for the population that is made up of the cattle of the smallholders, 0.3390±0.2030 for BLRE ex-situ population and 0.3894±0.1657 for the total TGC sample population (Tables II, III, IV ).
Some researchers like Özbeyaz et al. (1999), Altınalan (2005), Özkan (2005), Kurar et al. (2011) and Savaşçı and Atasoy (2016) reported the Ho average value in TGC they studied as 0.411±0.140, 0.433, 0.6823, 0.686 and 0.2788, respectively. Sharma et al. (2009) remarked that it varied between 0.70±0.10 and 0.65±0.14 in the 4 different Hind cattle race they examined and that the inadequacy of heterozygous cattle in the populations (Fis) was between 17.7 % and 28.8 %.
Table IV.- Results for the effective allele number (ne), the observed (Ho) and the expected (He) heterozygosity indices and the fixation index (Fis) tested in CAST and CAPN1 loci in the total TGC sample (n = 130).
| The loci |
ne |
1Ho |
1He |
2He |
3Fis |
| CAST |
1.9173 |
0.5615 |
0.4803 |
0.4784 |
- 0.1737*** |
| CAPN1 4751 |
1.9664 |
0.4692 |
0.4933 |
0.4914 |
0.0452ns |
| CAPN1 316 |
1.2472 |
0.1923 |
0.1990 |
0.1982 |
0.0297ns |
| Average |
1.7103 |
0.4077 |
0.3909 |
0.3894 |
|
| ± St. Deviation |
0.4018 |
0.1922 |
0.1663 |
0.1657 |
1, Levene (1949); 2, Nei (1987); 3, Wright (1965); ***, (P<0.001) significance; ns, non- significant.
When earlier the Turkish researchers tested the genetic differentiation in TGC population are compared to with the Ho average values (0.4077±0.1922) specified in my study, the values obtained in this study seems to be higher than the ones obtained by Savaşçı and Atasoy (2016), similar to the ones in Özbeyaz et al. (1999) study while they seem to be lower than the ones obtained in some other research. The cattle samples studied by Altınalan (2005), Özkan (2005), Kurar et al. (2011), Özşensoy and Kurar (2014) were also more heterogeneity.
The effective allele number and allele density are other criteria to show the spatial heterogeneity in a population. The average effective allele number per loci for TGC samples from the smallholders in the villages, BLRE TGC ex-situ conservation herd and the total population sample were calculated as 1.7379±0.3559, 1.6026±0.4422 and 1.7103±0.4018, respectively. CAPN1 316 loci (1.1245) in BLRE TGC ex-situ conservation herd seems to have the lowest effective allele number while CAST loci (1.992) in TGC in the villages was found to have the highest effective allele number. This shows that there is a linear relationship between Ho and ne. According to the results, it can be said that the herd under BLRE ex-situ conservation program is made up of more homogeneity cattle in terms of CAPN1 316 loci while TGC in the smallholders are more heterogeneity regarding CAST loci.
The significance control for the difference between the estimated average heterozygosity indices for the two subgroups (BLRE ex-situ and the smallholders) was held in t test table with respect to the probability α. While the expected heterozygosity difference between the two TGC subgroups for CAPN1 316 loci (P<0.001) and CAST loci (P<0.05) was found significant, the difference for CAPN1 4751 was not meaningful (P> 0.05).
CONCLUSION and Recommendations
Tables II, III and IV shows a linear relationship between the He values and ne. As is known, as ne in polymorphic loci draws closer to two, it is more likely to see heterogeneity in those loci. The more polymorphic the loci in a species or race are, the more the heterozygosity value will increase. In the total sample, it was found that the Ho (0.5615) and the He (0.4933) values were highest in CAST loci likewise ne value (1.9664) was highest in CAPN1 4751 loci (Table IV). When we look at the average ne, Ho and He values (Tables II and III) in subgroups, it was found that the sample TGC subgroups in the smallholders had the highest ne average (1.7379±0.3559), the highest He (0.4085±0.1377) and Ho (0.4430±0.1965) values. The difference between the average Ho value and the average He value in the total sample and BLRE ex-situ conservation herd being small implies the risk of increasing consanguinity and the beginning of a decrease in the genetic variation (Tables II and IV).
The low He average heterozygosity values of BLRE TGC ex-situ conservation herd can be attributed to the fact that are more closely related to the breeding system. Also the ne supports the increase in the homozygosity. Since the size of the active cattle population was small for breeding and the uncontrolled or faulty breeding programs could cause decrease in genetic variation in BLRE TGC ex-situ conservation herd. The excess in heterozygosity in CAST gene loci (27.09%) in the village group proves that the TGC in the villages are without any selection.
It can be said that there is a risk of decrease in the genetic variation in ex-situ conservation herd which is the result of the increasing consanguinity scaling up the homozygosis. Diminishing productivity, ability to transform feed into meat, and resistance to environmental conditions and illness, and increasing number of deformations are some of the most important negative effects of decreasing genetic variation as a result of increasing homozygosis (Karahan et al., 2007). Therefore, it is important to understand the genetic structures of animal populations to be able to protect the genotypic structure of the races that face the danger of becoming extinct.
According to the data obtained in this study, it can be suggested that cattle breeding herd from different herd be integrated into the herd that are under the ex-situ conservation program and the number of the cattle in the herd be increased. In this case, in this case it is thought that the sustainability of genetic variation will can provide.
Acknowledgement
This investigation has been supported by the project TUBAP-2013-109 accepted by Commission of Scientific Research Projects of Trakya University in Turkey.
Statement of conflict of interest
Authors have declared no conflict of interest.
References
Altınalan, A., 2005. Genetic characterization of microsatellite DNA markers in the indigenous cattle breeds in Turkey. PhD thesisi, Çukurova University, Institute of Science and Technology, Department of Animal Science. Adana, Turky, pp. 205.
Bozkurt, Y., 2012. Seasonal performance of different breeds of feedlot beef cattle grown under the Mediterranean conditions. Bulg. J. agric. Sci., 18: 443-445.
Hristov, P.I., Teofanova, D.R., Neov, B.S., Zagorchev, L.I. and Radoslavov, G.A., 2014. Population structure of two native bulgarian cattle breeds with regard to CSN3 and CSN1S1 gene polymorphism. Bulg. J. Vet. Med., 17: 18−24.
Karahan, B., Özcan-Gökçek, E. and Hekimoğlu, M., 2007. The effect of the genetic structure of the aquaculture and precautions to be taken. Aquacult. Fish., 8: 32-36.
Kımura, M. and Crow, J.M., 1978. Effect of overall phenotypic selection on genetic change at individual loci. Proc. natl. Acad. Sci., 75: 6168-6171. https://doi.org/10.1073/pnas.75.12.6168
Kök, S., Atalay, S., Eken H.S., Savaşçi, M., 2017. The Genetic Characterization of Turkish Grey Cattle with Regard to UoG CAST, CAPN1 316 and CAPN1 4751 Markers. Pakistan J. Zool., 49: 281-287. https://doi.org/10.17582/journal.pjz/2017.49.1.281.287
Kurar, E., Özsensoy, Y., Dogan, M., Bulut, Z., Altunok, V., Isık, A., Camlıdağ A. and Nizamlıoğlu, M., 2011. Autosomal and mitochondrial genetic diversity of Turkısh native cattle breeds. RBI 8th Global Conference on the Conservation of Animal Genetic Resources. Proceedings of Symposium, Tekirdağ, Turkey, pp. 179-184.
Levene, H., 1949. On a matching problem arising in genetics. Annls. Math. Stat. 20: 91-94. https://doi.org/10.1214/aoms/1177730093
Nei, M., 1987. Molecular evolutionary genetics. Columbia University Press, New York, pp. 505, ISBN-13: 978-0-231-06321-0.
Özbeyaz, C., Yıldız, M.A. and Çamdeviren, H., 1999. Genetic relationships among cattle breeds in Turkey. J. Lalahan Livest. Res. Inst., 39: 17-32.
Özkan, E., 2005. An investigation on genetic structure of native and cultural cattle breeds in Turkey by using microsatellite markers. PhD thesis, Institute of Science and Technology, Department of Animal Science, Trakya University, Tekirdağ,Turkey, pp. 235.
Özşensoy, Y., Kurar, E., Doğan, M., Bulut, Z, Altunok, V., Işık, A., Çamlıdağ, A., Nizamlıoğlu, M., 2010. Genetic characterization of some native cattle breeds in Turkey by using STR markers. Res. J. Biol. Sci. 3: 155-163. http://www.nobel.gen.tr/yayindetay.aspx?yayin_id=2081
Özşensoy, Y. and Kurar, E., 2014. Genetic diversity of native Turkish cattle breeds: Mantel, AMOVA and bottleneck analysis. J. Adv. Vet. Anim. Res., 3: 86-93. https://doi.org/10.5455/javar.2014.a21
Rincón, G. and Medrano, J.F., 2006. Assays for genotyping single nucleotide polymorphisms in the bovine CAPN1 gene. Anim. Genet., 37: 94–295. https://doi.org/10.1111/j.1365-2052.2006.01430.x
Sasimowski, E., 1987. Animal breeding and production. Science Publishers B.V. 0-444-99504-8., Elsevier, NL.
Savaşçı, M. and Atasoy F., 2016. The investigation of calpastatin and thyroglobulin gene polymorphisms in some native cattle breeds. Ankara Üniv. Vet. Fak. Derg., 63: 53-59. https://doi.org/10.1501/Vetfak_0000002709
Schenkel, F.S., Miller, S.P., Jiang, Z., Mandell, I.B., Ye, X., Li, H. and Wilton J.W., 2006. Association of a single nucleotide polymorphism in the calpastatin gene with carcass and meat quality traits of beef cattle. J. Anim. Sci., 84: 291−299. http://dx.doi.org/10.2527/2006.842291x
Sharma, R., Pandey, A.K., Singh, Y., Prakash, B., Mishra, B.P., Kathiravan, P., Singh, P.K. and Singh, G., 2009. Evaluation of genetic variation and phylogenetic relationship among North Indian cattle breeds. Asian-Aust. J. Anim. Sci., 22: 13-19.
Soysal, İ., Özder, M., Kök, S. and Soysal, D., 2005. Place in the ecosystem of Turkish grey cattle. 5th Industrialization and Environment Symposium in Thrace, TMMO Publishing, Edirne, Turkey, pp. 429-440.
Soysal, M.İ. and Kök, S., 2006. The last survivors of grey cattle’s whose resisting not to be extincted. A case study of characteristics and sustainability of traditional system of native grey cattle breed. 2nd Seminar of the scientific-Professional Network on Mediterranean Livestock Farming. Mediterranean Livestock Production: Uncertainties and Opportunities, CIHEAM Publishing, Zaragoza, ES, pp. 55-63.
Wright, S., 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution, 19: 395-420. https://doi.org/10.2307/2406450
Yeh, F.C., Yang, R., Boyle, T.J., Ye, Z. and Xiyan, J.M., 2000. POPGENE 32, Microsoft Window-based freeware for population genetic analysis, Version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, CA.
To share on other social networks, click on any share button. What are these?










