Development of 17 Novel Cross-Species Microsatellite Markers for Dabry’s Sturgeon (Acipenser dabryanus) from Chinese Sturgeon (Acipenser sinensis) via Next-Generation Sequencing
Development of 17 Novel Cross-Species Microsatellite Markers for Dabry’s Sturgeon (Acipenser dabryanus) from Chinese Sturgeon (Acipenser sinensis) via Next-Generation Sequencing
1Chinese Sturgeon Research Institute, China Three Gorges Corporation, Yichang Hubei 443100, China
2Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes, Yichang Hubei 443100, China
Yacheng Hu and Xueqing Liu contributed equally to this investigation and should be considered co-first authors.
ABSTRACT
In this study, we report 17 novel cross-species microsatellite markers for Dabry’s sturgeon via next-generation sequencing from Chinese sturgeon (Acipenser sinensis). The 17 microsatellites were polymorphic with 3 to 6 alleles per locus and the total number of alleles is 80. The mean expected heterozygosity (HE, 0.506 to 0.781), observed heterozygosity (HO, 0.542 to 1), Hardy-Weinberg departure value (d, -0.042 to 0.427), polymorphic information content (PIC, 0.482 to 0.753) and the Shannon-Wiener Diversity Indices (H’, 0.827 to 1.565) of all the 17 polymorphic loci indicated a high level of informativeness. Those markers will be useful tool to study the genetic diversity of Dabry’s sturgeon.
Article Information
Received 29 November 2018
Revised 13 February 2019
Accepted 27 March 2019
Available online 26 August 2019
Authors’ Contributions
YH, HD and XL conceived and designed the research. YH wrote the manuscript with contributions from HD and JY. BW analyzed the data. YH, KX and XL carried out the experiment.
Key words
Microsatellite, Acipenser dabryanus, Genetic diversity, Development, Cross-species.
DOI: http://dx.doi.org/10.17582/journal.pjz/2019.51.6.sc4
* Corresponding author: [email protected]
0030-9923/2019/0006-2381 $ 9.00/0
Copyright 2019 Zoological Society of Pakistan
Dabry’s sturgeon (Acipenser dabryanus), inhabited the upper and middle sections of the Yangtze River, is a kind of endemic species in China. Dabry’s sturgeon was commercial fisheries before the 1980s. However, the number of Dabry’s sturgeon wild population declined drastically due to environmental pollution, such as overfishing, and habitat alteration and destruction (Zhuang et al., 1997). Nowadays, Dabry’s sturgeon is listed as Critical Endangered species in the Red List of Threatened Species (http://www.iucnredlist.org). A lot of efforts, including artificial releasing, re-stocking of wild juveniles, controlled reproduction and national nature reserve, have been done to save this species (Zhang et al., 2011). Nevertheless, the status of Dabry’s sturgeon in the Yangtze River are still further worsened as habitat degradations and poaching. Therefore, Dabry’s sturgeon has been intensively breeding in artificial condition for conservation purposes.
Dabry’s sturgeon and Chinese sturgeon Acipenser sinensis are only two species of acipenserids found in the Yangtze River, China. The two species are so similar that they are not scientifically distinguished until the middle of the 19th century. There was some taxonomic confusion between Chinese sturgeon and Dabry’s sturgeon until the 1960s. Species identification of Chinese sturgeon and Dabry’s sturgeon is mainly based in body color, gill raker number, skin roughness between ganoid scales and length. It is accept that Chinese sturgeon is an anadromous species that inhabits the Yangtze River, the East China and Yellow Seas, while Dabry’s sturgeon is a freshwater species restricted to the Yangtze River (Zhuang et al., 1997). Dabry’s sturgeon has been characterized as a landlocked ecotype of Chinese sturgeon in the Yangtze River basin.
Microsatellite plays an important role in population genetics research, and has been of great utility in conservation and management of biological resources (Chistiakov et al., 2006). To better address the conservation status of Dabry’s sturgeon in the Yangtze River, a management project based on genetic is being used to define the efficiency of artificial releasing. Development microsatellite markers are helpful to make a protective strategy of Dabry’s sturgeon (Zhu et al., 2005). A lot of microsatellite markers have been developed for Dabry’s sturgeon in previous studies (Que et al., 2014; Zeng et al., 2013; Zhang et al., 2013). However the loci in previous studies only provided little data about genetic analysis, especially polymorphic information content (PIC) of microsatellites markers. Besides, the traditional method of the microsatellite marker developing was time-consuming, costly, laborious and often inefficient (Zane et al., 2002). Microsatellite is conserved among closely related species and may be useful to clarify interspecific genetic relationships between recently diverged taxa (Dawson et al., 2010). However, the microsatellite developed for one species may not always be useful for others because of their high mutation rate. The mutations at primer binding sites may lead to disruptions within tandemly repeated elements that can reduce the level of observed polymorphism in the non-target species. The development of microsatellite should be closely related to maximize utility and offset the costs. Therefore, it has become fairly standard practice to test the newly developed microsatellite loci for cross-species utility (Huang et al., 2014; Wang et al., 2018). In this study, we report 17 novel cross-species microsatellite markers for Dabry’s sturgeon from Chinese sturgeon via next-generation sequencing, and those markers will be useful for the research of genetic management, genetic analysis and supportive breeding evaluation.
Table I.- Characterization of 17 microsatellite loci in Dabry’s sturgeon.
|
Loci |
GenBank accession No. |
Repeat motif(s) |
Primer sequence (5'-3') |
Tm (°C) |
|
ZHX70 |
MH667327 |
(TATC)11 |
F:GGCATTATAGACCCCTGTCGG |
60 |
|
R:ACAGCTGGGGAGGAACAGTA |
||||
|
ZHX80 |
MH667328 |
(ATAA)10 |
F:AGTGTTGTGAATAGGACCCAAAGT |
60 |
|
R:GCATTTTGCTGTACAGACCTTGA |
||||
|
ZHX91 |
MH667329 |
(TTTA)10 |
F:TTGGCGATCCGATCACCAAA |
60 |
|
R:TGCCATTTGACTCAACTGTGC |
||||
|
ZHX76 |
MH667330 |
(TATC)14 |
F:GCGTTCACTGAGTCAATGCA |
59 |
|
R:CTGGACAGAGAACAGATAGCGT |
||||
|
Z15 |
MH667331 |
(ATAG)14 |
F:AGCTAGCAACTGAAGCCCTG |
60 |
|
R:ACAGCTGCAGCACACTTTTG |
||||
|
Z120 |
MH667332 |
(TCTT)13 |
F:CTGTGTCTTCTGCTCCTGGT |
59 |
|
R:GCATGTCAGGGCCGGTATTA |
||||
|
Z125 |
MH667333 |
(AGAT)12 |
F:CTGGCTAAGGTCATGGCCAA |
58 |
|
R:CAAACTTTATTTGGAAAATTGCACAAT |
||||
|
Z153 |
MH667334 |
(TCTT)13 |
F:TGGACTGACAACACTGCTCC |
60 |
|
R:ACAGAGCACGTACAGCCAAA |
||||
|
Z166 |
MH667335 |
(ATCT)15 |
F:GTGTTTAGAATGTTTAAACACTGAGGA |
60 |
|
R:TGGCCTCAAGTTCAAGCACA |
||||
|
Z167 |
MH667336 |
(GAAA)13 |
F:GCACGAGAGAGACAGGACAA |
59 |
|
R:CAGGTTGAAAGTGCTGGTGC |
||||
|
Z175 |
MH667337 |
(AGAA)12 |
F:GACGCGCTCTCTGCAATTTC |
59 |
|
R:TCTCACCTCAATTCTCGTGAGT |
||||
|
Z181 |
MH667338 |
(ACTA)10 |
F:TTCGGTAACAAGACGCTGCT |
58 |
|
R:AGGGAAATACCTGTACATTTAAGAGA |
||||
|
Z182 |
MH667339 |
(AGAT)10 |
F:TGCAACTGGGTTCACTATGCA |
60 |
|
R:GATCATGCCCTGGCTTCTGA |
||||
|
Z189 |
MH667340 |
(TGTC)10 |
F:TGGTTCCTCCCAGTCCTCAA |
60 |
|
R:GTGAGGAGCAGACTGGACTG |
||||
|
Z193 |
MH667341 |
(TCTT)8 |
F:GGGCCGTTCTGCTTTTTGAG |
60 |
|
R:CCCCGTGTAGGACACTAACG |
||||
|
Z199 |
MH667342 |
(TCTT)13 |
F:GACGTTTGGAGCGTGGAAAC |
60 |
|
R:TGGCATTTACAGCATAAACTAAACCT |
||||
|
Z219 |
MH667343 |
(AAAG)9 |
F:GTCCAGGGCAGCATCTACTG |
60 |
|
R:CTCAAACGCACAACCAGCAA |
Tm, annealing temperature.
Materials and methods
The RNA was extracted from the fin of Chinese sturgeon using Trizol (Invitrogen, USA), according to the manufacturer protocol. RNase-free DNase I was used to remove DNA from the RNA and the total RNA was evaluated by 1% agarose gel electrophoresis. The cDNA library was created by using approximately 5 μg RNA and was sequenced on an Illumina Hiseq2000. The sequences were selected by constrained to perfect repeat motifs of 4 bp from the cDNA library by the Microsatellite Identification tool (MISA: http://pgrc.ipkgatersleben.de/misa/). All the selected sequences were then used to design microsatellite primers using Primer Premier 5.0 software (http://www.premierbiosoft.com/primer design/).
We collected 24 individuals of Dabry’s sturgeon from the Chinese Sturgeon Research Institute in Yichang City, China. The total DNA was extracted from the fin of Dabry’s sturgeon by the method of rapid salt-extraction (Aljanabi and Martinez, 1997). Polymerase chain reaction (PCR) was carried out using the DNA as template. The recommended cycling conditions of PCR is 3 min at 94°C; 35 (30s at 94°C, 30s at annealing temperature; 30s at 72°C); 10 min at 72C. The PCR was carried out in a 25-ul reaction volume, containing 0.25μM for each primer, 1×PCR buffer (TaKaRa), 1.5 mM MgCl2, 50-100 ng genomic DNA and 0.25 U Taq DNA polymerase (TaKaRa). The PCR products were visualized on 10% polyacrylamide gel stained (PAGE gel) with silver staining. The pBR332 DNA/MspI molecular marker (TaKaRa) was used as a standard to identify size of alleles.
The statistics of the observed heterozygosity (HO), the mean expected heterozygosity (HE) and Shannon-Weiner Diversity Indices (H’) for every loci were performed using software ATetra1.2 (Puyvelde et al., 2010). The Hardy-Weinberg departure value (d) and the polymorphic information content (PIC) were performed according to formula:
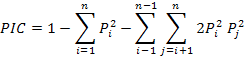
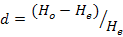
Where, Pi and Pj is the frequency of I and J allele in the group.
All fish handling and experimental procedures in this study were approved by the Chinese Sturgeon Research Institute, the China Three Gorges Corporation, and the Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes.
Results
In this study, a total of 96 sequences were found and were selected to designing microsatellite primer. The 96 sequences that tested in Dabry’s sturgeon were consistent with Chinese sturgeon. Among the 96 pairs of microsatellites primers, 27 pairs failed to produce any PCR product, 52 pairs were produced low quality results, and 17 pairs showed polymorphism (Table I). The percentage of amplification success was 17.7%. The percentage of errors by replicating 5 samples was 0. Using the 17 microsatellite markers, we can identify each individuals of all the 24 Dabry’s sturgeon. The 17 microsatellites sequences were polymorphic with 3 to 6 alleles per locus and the total number of alleles is 80. The mean expected heterozygosity (HE) and observed heterozygosity (HO) ranged from 0.506 to 0.781, from 0.542 to 1, respectively. The Hardy-Weinberg departure value (d, -0.042 to 0.427), polymorphic information content (PIC, 0.482 to 0.753) and the Shannon-Wiener Diversity Indices (H’, 0.827 to1.565) of all the 17 polymorphic loci indicated a high level of informativeness (Table II).
Table II.- Genetic diversity for 17 microsatellite markers in Dabry’s sturgeon.
|
Loci |
HE |
HO |
d |
PIC |
H' |
Na |
Allele range |
|
ZHX70 |
0.701 |
1 |
0.427 |
0.653 |
1.266 |
4 |
220-252 |
|
ZHX80 |
0.781 |
1 |
0.28 |
0.753 |
1.565 |
5 |
173-217 |
|
ZHX91 |
0.772 |
0.958 |
0.241 |
0.75 |
1.542 |
5 |
202-242 |
|
ZHX76 |
0.746 |
1 |
0.34 |
0.724 |
1.494 |
6 |
224-300 |
|
Z15 |
0.743 |
1 |
0.346 |
0.725 |
1.505 |
6 |
206-238 |
|
Z120 |
0.781 |
1 |
0.28 |
0.749 |
1.562 |
5 |
160-188 |
|
Z125 |
0.705 |
0.792 |
0.123 |
0.672 |
1.274 |
4 |
240-272 |
|
Z153 |
0.724 |
1 |
0.381 |
0.676 |
1.331 |
4 |
222-250 |
|
Z166 |
0.774 |
1 |
0.292 |
0.745 |
1.545 |
5 |
232-260 |
|
Z167 |
0.609 |
0.583 |
-0.042 |
0.532 |
1 |
4 |
218-262 |
|
Z175 |
0.726 |
1 |
0.377 |
0.683 |
1.333 |
4 |
186-226 |
|
Z181 |
0.722 |
1 |
0.385 |
0.683 |
1.328 |
4 |
190-246 |
|
Z182 |
0.768 |
1 |
0.302 |
0.739 |
1.529 |
5 |
172-200 |
|
Z189 |
0.709 |
0.917 |
0.293 |
0.666 |
1.347 |
5 |
250-310 |
|
Z193 |
0.722 |
0.958 |
0.327 |
0.698 |
1.392 |
5 |
170-210 |
|
Z199 |
0.681 |
0.917 |
0.346 |
0.664 |
1.34 |
6 |
225-253 |
|
Z219 |
0.506 |
0.542 |
0.07 |
0.482 |
0.827 |
3 |
200-280 |
HE, mean expected heterozygosity; HO, observed heterozygosity; d, Hardy-Weinberg departure value; PIC, polymorphic information content; H’, Shannon–Wiener Diversity Indices; Na, number of alleles in per locus.
Discussion
In this study, 17 novel cross-species microsatellite markers were developed for Dabry’s sturgeon from Chinese sturgeon via next-generation sequencing for the first time. All the 17 microsatellite markers showed a high polymorphism (PIC>0.5), except Z219. No microsatellite marker showed significant deviation from the Hardy-Weinberg departure value (d < 0.05) except Z167, indicating that they were polymorphism. Little genetic analysis was performed in previous microsatellites studies of Dabry’s sturgeon (Que et al., 2014; Zeng et al., 2013; Zhang et al., 2013). For example, the calculation of HO, d and PIC were totally or partially missing in these studies. Considering that HO, d and PIC are indicative of population genetic variation, microsatellite markers without those information are hard to determine whether they are suitable for population analysis. The high estimates of PIC of the microsatellites in this study substantiated the suitability of the markers to applications such as genetic diversity study, linkage-mapping programs in Dabry’s sturgeon. Compared to traditional method, we developed a cheaper and faster way to identify microsatellite loci. This method can be used as a reference for relative studies in other species.
The number of alleles per microsatellite locus developed in Que et al. (2014) study, Zeng et al. (2013) study, Zhang et al. (2013) study is 1 to 11, 4 to 14 and 3 to 13, respectively. The number of alleles per microsatellite locus in this study was less than the previous studies, with the value of 3 to 6. Maybe the samples used in this study were consisted sib individuals, or the microsatellites were cross-amplified. The mean expected heterozygosity of microsatellites in this study (0.506 to 0.781) is similar to the microsatellites developed by Zeng et al. (2013) (0.487 to 0.874) and is better than the microsatellites developed by Que et al. (2014) (0.127 to 0.828) and Zhang et al. (2013) (0.303 to 0.806). The mean expected heterozygosity in this study may be higher than the fact due to the fact that the number of alleles per microsatellite locus in this study is less than others. Only 17 out of 96 microsatellites showed polymorphism, and the rest microsatellites produced low quality results or even nothing. We sequenced all the microsatellites in this study by Dabry’s sturgeon DNA. Although those microsatellites were developed based on cDNA sequences of Chinese sturgeon, they can also be used to analysis the genetic of Dabry’s sturgeon, proving that the relationship between Chinese sturgeon and Dabry’s sturgeon is very close. In this study, all loci in each sample have no more than four alleles, which is consistent with two previous studies (Que et al., 2014; Zeng et al., 2013). Those microsatellite markers in this paper will be useful tool to study the population structure and genetic diversity of Dabry’s sturgeon.
This work was supported by Three Gorges Environment Protection fund, Chinese Three Gorges Corporation (No: XN270).
The experiments were performed in accordance with relevant institutional and national guidelines and regulations for the care and use of laboratory animals.
Statement of conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this article.
References
Aljanabi, S.M. and Martinez, I., 1997. Nucl. Acids Res., 25: 4692-4693. https://doi.org/10.1093/nar/25.22.4692
Chistiakov, D.A., Hellemans, B. and Volckaert, F.A.M., 2006. Aquaculture, 255: 1-29. https://doi.org/10.1016/j.aquaculture.2005.11.031
Dawson, D.A., Horsburgh, G.J., Küpper, C., Stewart, I.R.K., Ball, A.D., Durrant, K.L., Hansson, B., Bacon, I., Bird, S., Klein, Á. Krupa, A., Lee, J.W., Simeni, M., Smith, G., Spurgin, L. and Burke, T., 2010. Mol. Ecol. Resour., 10: 475-494. https://doi.org/10.1111/j.1755-0998.2009.02775.x
Huang, D., Zhang, Y., Jin, M., Li, H., Song, Z., Wang, Y. and Chen, J., 2013. Mol. Ecol. Resour., 14: 569-577. https://doi.org/10.1111/1755-0998.12197
Puyvelde, K.V., Geert, A.V. and Triest, L., 2010. Mol. Ecol. Resour., 10: 331-334. https://doi.org/10.1111/j.1755-0998.2009.02748.x
Que, Y., Xu, D., Shao, K., Xu, N., Li, W. and Zhu, B., 2014. J. Genet., 93: e62-65. https://doi.org/10.1007/s12041-014-0363-2
Wang, L., Gong, Y., Chen, K., Wang, H. and Lyu, X., 2018. Pakistan J. Zool., 50: 791-793. http://dx.doi.org/10.17582/journal.pjz/2018.50.2.sc8
Zane, L., Bargelloni, L. and Patarnello, T., 2002. Mol. Ecol., 11: 1-16. https://doi.org/10.1046/j.0962-1083.2001.01418.x
Zeng, Q., Ye, H., Ludwig, A., Wang, Z., Zhang, Y. and Peng, Z., 2013. J. appl. Ichthyol., 29: 1219-1221. https://doi.org/10.1111/jai.12278
Zhang, H., Wei, Q.W., Du, H. and Li, L.X., 2011. J. appl. Ichthyol., 27: 181-185. https://doi.org/10.1111/j.1439-0426.2011.01674.x
Zhang, S.H., Luo, H., Du, H., Wang, D.Q. and Wei, Q.W., 2013. Conserv. Genet. Resour., 5: 409-412. https://doi.org/10.1007/s12686-012-9815-2
Zhu, B., Liao, X., Shao, Z., Rosenthal, H. and Chang, J.B., 2005. Mol. Ecol. Resour., 5: 888-892. https://doi.org/10.1111/j.1471-8286.2005.01100.x
Zhuang, P., Ke, F.E., Wei, Q., He, X. and Cen, Y., 1997. Environ. Biol. Fishes, 48: 257-264. https://doi.org/10.1023/A:1007399729080
To share on other social networks, click on any share button. What are these?










