Isolation and Molecular Characterization of Bovine Coronavirus from Calves with Acute Gastroenteritis in Egypt
Isolation and Molecular Characterization of Bovine Coronavirus from Calves with Acute Gastroenteritis in Egypt
Saad A. Moussa1, Ahmed F. Afify1*, Suzan Salah2 and Ayman Hamed3
Electrophoresis of the amplified PCR product of BCoV N-gene (M.W band at 236 bp), first Lane (L) is 100bp DNA standard ladder, the second lane is a positive control, the third one is a negative control, and lanes from 1-4 are positive samples.
Phylogenetic tree for the partial nucleotide sequence of BCoV N-gene (BCoV_AHRI_EGY_2020- Acc. No. MW173144, Red circled) compared with other reference strains in the Genbank database. The scale bar represents the number of substitutions per nucleotide.
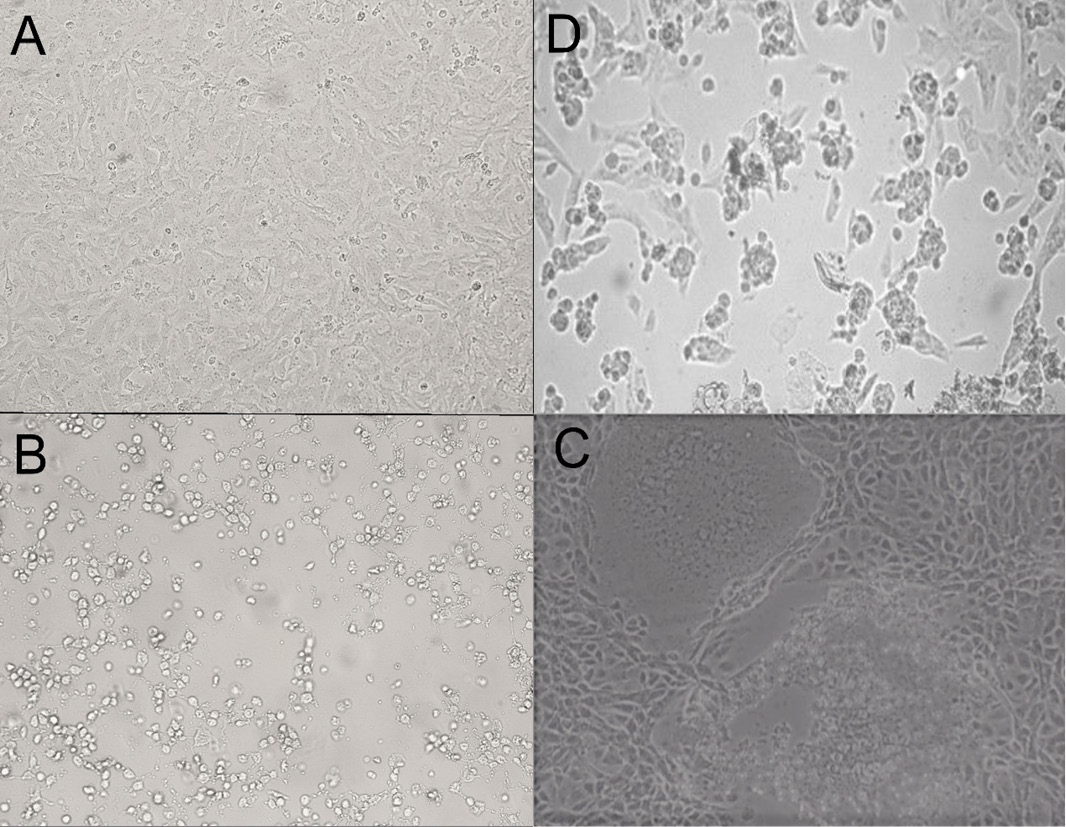
Microscopic pictures for BCoV CPE in Vero cells (3rd passage) at 100X magnification power; image A: control uninfected Vero cells, image B: rounding of cells 24 H.P.I, granulation, image C: swelling and syncytium formation 96 H.P.I., image D: cell lysis and detachment 5 days post-inoculation.
Image of BCoV visualized by TEM (negative staining) showing characteristic radial projections forming a crown shape like pointed by the black arrow, with an average of 140nm diameter confirming identity of BCoV isolate.
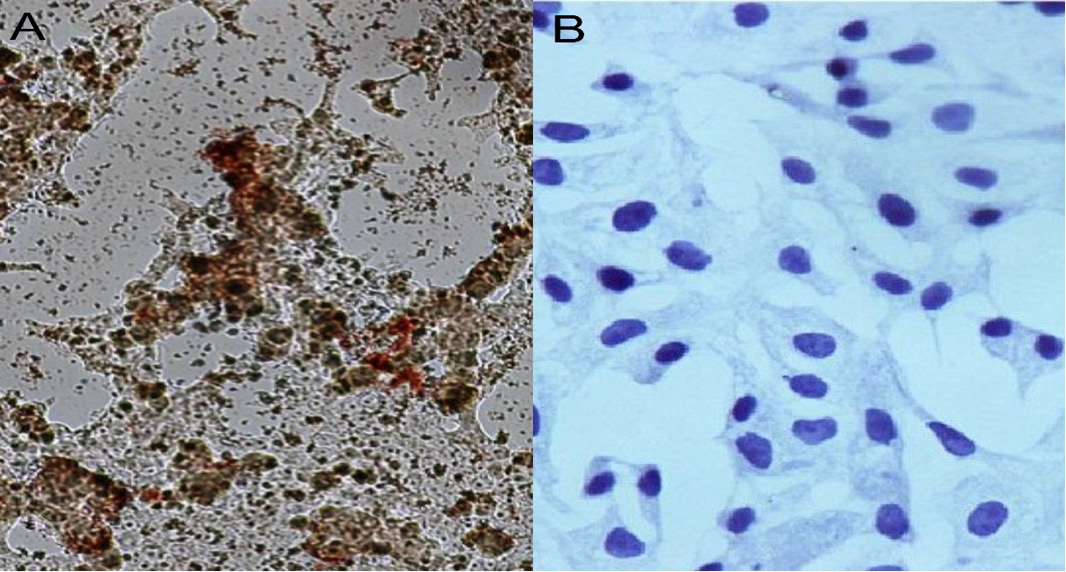
The image of IP test on BCoV isolate in Vero cells 18 H.P.I, image A: tested sample at 200X MP showed a high average of brownish stained cells confirming virus identity, where image B: negative control cells at 400X MP were not stained.
Percentage of nucleotide sequence identity between BCoV_AHRI_EGY_2020 strain and BCoV reference strains published on Genbank based on partial sequence of N-gene.
Percentage of amino acids sequence identity between BCoV_AHRI_EGY_2020 strain and BCoV reference strains published on Genbank based on partial sequence of N-gene.