Effect of Metformin on P53 and its Interacting Partners in Breast Cancer Cell Line MCF-7
Effect of Metformin on P53 and its Interacting Partners in Breast Cancer Cell Line MCF-7
Mehroze Amin1, Afifa Yaqub1, Qindeel Fatima1, Rabail Hassan Toor1,3 and Abdul Rauf Shakoori1,2*
1School of Biological Sciences, University of the Punjab, Quaid-i-Azam Campus, Lahore 54590, Pakistan
2Cancer Research Centre, University of the Punjab, Quaid-i-Azam Campus, Lahore 54590, Pakistan
3UVM Cancer Center, Larner College of Medicine, Department of Biochemistry, University of Vermont, Burlington, VT 05405, USA
ABSTRACT
Metformin is known to favor p53 anti-cancer activity and promote apoptosis and cell senescence in cancer cells. It is also known to decrease the incidence of several cancers though it is not entirely true for breast cancer. In the present study, the effect of metformin has been studied on the expression of p53 and its interacting partners MDM-2, PIRH2 and ΔNp73 in breast cancer cell line MCF-7 and non-cancerous kidney cell line HEK2093. Analysis of the cytotoxicity and proliferation parameters showed that higher concentrations of metformin were cytotoxic for the breast cancer cells with EC50 of 22.75µM. Analysis of expression of p53 and the interacting partner genes showed that metformin upregulated p53 expression in a dose-dependent manner, while the expression of interacting partners is downregulated indicating that metformin exhibits the anti-cancerous effect by regulating the p53 pathway. Altogether this study reports the effect of metformin on the expression of p53 and its interacting partners MDM2, PIRH2 and ΔNp73in breast cancer cell line MCF-7, thereby providing evidence that metformin modulates p53 pathway to exhibit its anti-cancer property. Hence , metformin can be a potential therapeutic candidate for breast cancer treatment. However, further investigations are required to provide insight into the therapeutic potential od metformin for cancer.
Article Information
Received 02 January 2022
Revised 15 February 2022
Accepted 02 March 2022
Available online 30 March 2022
(early access)
Published 23 August 2022
Authors’ Contribution
ARS conceptualization, fund acquisition, project administration, supervision, writing, review, editing and provided resources. MA methodology, writing original draft. AY help in methodology. QF methodology help in cell culture. RHT writing, review and editing.
Key words
Breast cancer, MDM2, PIRH2, ΔNP73, Metformin, Metastasis, P53
DOI: https://dx.doi.org/10.17582/journal.pjz/20220102211525
* Corresponding author: [email protected], [email protected]
0030-9923/2022/0006-2699 $ 9.00/0
Copyright 2022 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Breast cancer is one of the most lethal and diversified malignant tumors in the world. It is a general health dilemma for women and accounts for 24.2% of female cancers up to 2018 (Akram et al., 2017). Cancer recurrence and metastasis create problems that lead to a high mortality rate. There is a critical need for discovering unique target genes for the prediction and treatment of breast cancer (Zhang et al., 2021). P53 also known as guardian of human genome induces senescence, growth arrest, and apoptosis to play the role of a tumor suppressor gene. In 50% of human cancers, it is known to be suppressed or inactivated by mutations. As mutant p53 is frequently found involved in cancer proliferation it is known to be an exciting and promising target in cancer therapy (Wang and Sun, 2010).
Metformin, an anti-diabetic drug that is recently being studied for its anti-cancer properties, works with the p53 gene to exert its anti-tumor effects. Metformin limits oxidative phosphorylation that results in reduced ATP formation and ultimately AMP level in the cell rises. AMP-activated protein kinase (AMPK) phosphorylates the p53 gene as a result p53 attains stabilized structure and becomes activated. P53 being a tumor suppressor gene plays a major role in apoptosis and cellular senescence of the cancer cells. P53 acts in a positive feedback loop with AMPK by inhibiting mTOR (mammalian target of rapamycin) to arrest cell progression (Faria et al., 2019). Isleem et al. (2020) showed that metformin combined with olive leaf extract resulted in reduced viability in the case of cancer cells through a powerful synergistic inhibitory effect.
Protein levels of p53 and its transcriptional activity are tightly regulated by E3 ubiquitin Mdm2–protein ligase Mdm2, encoded by MDM2 gene, the expression of which is transcriptionally regulated by p53 in a negative feedback loop (GarcíaCano et al., 2020). In 2021, studies were carried out to find the effect of 3, 4-dihydroxy-5, 4′-dimethoxybibenzyl (DS-1) on targeting MDM2 and restoring p53 function in lung cancer cells, as targeting MDM2, a negative regulator of p53 has recently attracted interest in cancer drug research as it may restore tumor suppressive function (Putri et al., 2021).
The oncogenic ΔNp73 controls cell survival and self-renewing properties of stem cells, and promotes increased susceptibility to carcinogenesis. ΔNp73 overexpression has been commonly associated with lymph node metastases and vascular invasion, chemoresistance, and poorer patient prognosis. ΔNp73 also acts as a negative regulator of P53 thereby sustaining cancer cell proliferation (Ramos et al., 2020).
Pirh2, a ubiquitin-protein ligase, has been reported to promote p53 degradation. Pirh2 overexpression could potentially cause degradation of p53 and reduce its tumor suppression function in the lung tumor cells (Duan et al., 2006).
The aim of this study was to determine the effect of metformin on the expression level of the p53 gene and its interacting partners (PIRH2, MDM-2, and ΔNp-73) in breast cancer cell line MCF-7 and normal human embryonic kidney HEK-293 cell line. This study aimed to determine the anti-cancer effect of metformin concerning the p53 pathway.
Materials and Methods
Maintenance of cell lines
In this study, breast cancer cell line MCF-7, and normal embryonic kidney cell line HEK-293 (CRL-1573TM) acquired from ATCC (American Type Culture Collection, Manassis, VA, USA (catalog No. 11686-029 and 30-2003, respectively) were used. The cell cultures were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Catalogue No. 11995065, GIBCO, Thermofisher Scientific, Waltham, MA, USA) with 10% heat-inactivated fetal bovine serum (FBS, Catalogue No. 10082147, GIBCO) and 1 % streptomycin/penicillin (catalog No. 15140148, GIBCO). Cells were maintained at 37°C in a humid atmosphere with 5% CO2 maintained in air.
In vitro cytotoxicity assay
MCF-7 cells were seeded at the density of (3.0×103 cells per cm2) in a 24-well culture plate. After 24 h of seeding, metformin treatment was given in a series of concentrations (1 µM, 5 µM, 10 µM, 20 µM, 30 µM). Untreated (control) cells were treated with a complete growth medium supplemented with 10 µl 1X PBS.
After 48 h of metformin treatment, neutral red assay was performed. The culture plates were incubated for 2 h in a CO2 incubator following the procedure mentioned in Repetto et al. (2008). The dye was extracted and its optical density was measured at 570 nm by using BioTek-808 ELISA absorbance reader. Since the assay is based on the lysosomal uptake of dye, the absorbance corresponded to the number of viable cells.
Percentage cell viability was calculated by using the following formula:
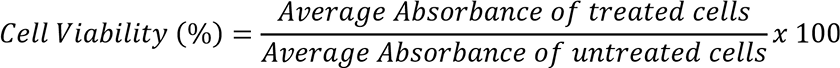
Growth inhibition and EC50 were calculated by using Microsoft Excel.
In vitro cell proliferation assay
BrdU assay was carried out following the protocol provided by Sigma Aldrich (Sigma BrdU Cell Peroliferation ELISA Kit Catalog no. 11296736001). Briefly, 1X BrdU reagent was added to each well for 4 h at 37 °C and 5% CO2. Cells were fixed and DNA was denatured to make incorporated BrdU dye more accessible for detection by antibodies. The pre-diluted anti-BrdU primary or secondary peroxidase-conjugated monoclonal detector antibodies were added to each well (100 µl/well) and incubated for 1 h at room temperature. After washing thrice with washing buffer, the bound peroxidase was detected by substrate reaction, which was stopped by adding stop solution (100 µl/well), and then O.D of the yellow reaction product was measured at 450 nm using a BioTek-808 Elisa absorbance reader. The intensity of the color was proportional to the BrdU incorporation in the DNA. For metformin-treated cells, the percentage cell proliferation was calculated using Microsoft Excel taking untreated cells (control) as 100%.
The following formula was used to calculate percentage cell proliferation.
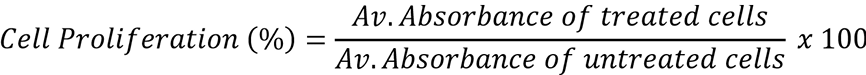
Quantitative real-time RT-PCR (qRT-PCR)
After 48 h of the treatment of metformin with a series of concentrations of metformin (1 µM, 5 µM, 10 µM, 20 µM, 30 µM) total RNA was isolated from treated and untreated cells using the TRIzol method. cDNA synthesis Kit (RevertAid First Strand cDNA synthesis kit-(catalog number: K1621) was used for cDNA synthesis. Primers used in this study are shown in Table I.
Each cDNA template was diluted to make the final concentration 40 ng/ µl. Maxima SYBR Green/ Low ROX qPCR Master Mix (catalog no. K0221) was used to set up the experiment of RT PCR where untreated samples were used as a calibrator and HMBS was used as a reference gene for normalization. The reaction was carried out in triplicates (biological replicates). Comparative gene expression was checked using the 2–∆∆Ct method.
Statistical analysis
The graphs were plotted using Microsoft Excel and GraphPad Prism (v 7.03). Statistical analyses were carried out by using GraphPad Prism (V.7.03). For the cell-based assays, one-way ANOVA with Dunnett’s test for multiple comparisons was carried out while for the real-time PCR, the statistical analysis was performed using two-way ANOVA with Tukey’s test for multiple comparisons. In the case of both tests, analysis was performed with a 95% confidence interval with statistical significance at P≤0.05. Further indication of the level of statistical significance is shown as follows: *(p ≤0.05), **(p≤0.01), ***(p≤0.005), ****(p≤0.001).
Table I. Primers for quantitative (real-time) PCR of target specific genes used in this study.
|
Target gene |
Primer sequence 5’ 3’ |
Product size |
Melting temperature ™ |
|
PIRH2 |
TGGGGTAAGTGTTGTCCTCA |
137bp |
58.9°C |
|
TGTGGTAAAAGTGGGTTTGG |
58.3°C |
||
|
ΔNp73 |
CAGCCCATCAAGGAGGAGTT |
127bp |
61.5°C |
|
TGAGGCAGTTTTGGACACAC |
59.7°C |
||
|
MDM2 |
TGCCATTGAACCTTGTGTGA |
127bp |
60°C |
|
GCAGGGCTTATTCCTTTTCT |
58°C |
||
|
P53 |
AGAGTCTATAGGCCCACCCC |
140bp |
60.04°C |
|
GCTCGACGCTAGGATCTGAC |
59.96°C |
||
|
HMBS |
TACCCCGAGAGGAGAGAACA |
203bp |
59°C |
|
CGAGCAGGAAGACCAGAAAC |
60°C |
*All primers were designed using Primer-Blast by NCBI (https://www.ncbi.nlm.nih.gov/).
RESULTS
Morphology of cells
The HEK-293 human embryonic kidney cell line was used as a model for non-cancerous cells in this study. Both MCF-7 and HEK-293 cell lines have epithelial-like morphology (Fig. 1). Morphologically it was observed that the untreated cells (control) and the cells treated with lower concentrations of metformin i.e., 1 µM and 5 µM exhibited their normal morphology, while the cells treated with higher metformin concentrations i.e., 10 µM, 20 µM, and 30 µM exhibited stressed morphology, with smaller sizes and lesser number of cells compared to control cells (Fig. 2).
Cell viability and cell proliferation
Cell viability of MCF-7 cells treated with different concentrations of metformin was determined by lysosomal uptake of neutral red dye. Figure 3A shows that the cells treated with 1 µM metformin had comparable cell viability (97.8 %) to the control cells. The cell viability was significantly (P≤0.05) reduced in cells treated with 5 µM metformin with 82.2 % viable cells. At higher concentrations, further reduction in cell viability was observed with 72 %, 69.7 %, and 33.9 % viable cells at 10 µM, 20 µM, and 30 µM metformin-treated cells respectively. We observed a dose-dependent relation of metformin on cell viability.
BrdU, a synthetic thymidine analog, incorporation into DNA was determined to determine cell proliferation. Figure 3B shows the effect of metformin on the proliferation of MCF-7 cells. We observed a dose-dependent reduction in cell viability with increasing metformin concentrations in the MCF-7 cell line. The analysis of the percentage cell proliferation showed that the concentrations 1 µM, 5 µM, and 10 µM had 86.4 %, 85.2 %, and 79.1 % proliferative cells respectively, which although was less than the control, the difference was not found to be statistically significant with P≥0.05. However, at higher concentrations of metformin i.e., 20 µM and 30 µM the cell proliferation was found to be 64 % and 53.9 % which was significantly reduced (P≤0.05) compared to the control (Fig. 3B).
Figure 4 shows the effective concentration of metformin with 50 % cell viability of MCF-7 cells (EC50=22.75 µM) based on cell viability data.
According to the outcomes, metformin demonstrated a concentration-dependent anti-proliferative activity in MCF-7 cells.
Effect of metformin on p53 gene expression
Analysis of the expression of the p53 gene showed that in experimental controls of both MCF-7 and HEK cells, the expression of p53 was at the basal level. However, metformin treatment to MCF-7 upregulated the expression of the p53 gene in a concentration-dependent manner. In MCF7 cells, the p53 expression was upregulated 1.8-fold, 2.26-fold, and 3.20-fold after treatment with 1, 5, and 10 µM metformin. This increase was 4.3-fold and 8.8-fold, respectively after treatment with 20 µM and 30 µM metformin (Fig. 5A). A similar trend in the expression of p53 was observed in metformin-treated HEK-293 cells. The cells at 1 µM, 5 µM, and 10 µM had 1.16-fold, 1.35-fold, and 1.31-fold p53 expression respectively, which was upregulated to 12.8-fold and 11.47-fold in 20 µM and 30 µM metformin-treated HEK-293 cells, respectively.
Effect of metformin on the expression of interacting partner genes of p53 gene
Figure 5 shows the effect of different concentrations of metformin chloride on the interacting partner genes such as ΔNp73, PIRH2, MDM-2 in MCF-7 and HEK-293 cell lines. The relative expression of ΔNp73 was 10.79 in MCF7 and 7.37 in HEK-293 experimental controls. Compared to the experimental control, the expression of the ΔNp73 gene was significantly (P≤0.05) downregulated in metformin-treated MCF-7 cells in a dose-dependent manner. The relative expression recorded was 4.16, 3.85-fold, 2.27, 2.2, and 1.41 after treatment of MCF-7 cells with 1, 5, 10, 20, and 30 µM metformin (Fig. 5B). HEK-293 cells also showed statistically significant (P≤0.05) downregulation in the expression of the ΔNp73 gene compared to its experimental control, except for 1 µM metformin treatment), where the difference in expression was not significant compared to the experimental control sample.
Pirh2 is another interacting partner and negative regulator of p53. In MCF-7 cells, the relative expression of PIRH2 was 8.17 in the experimental control sample, which was downregulated after metformin treatment (Fig. 5C). An opposite trend was observed in the metformin-treated HEK-293 cells, where a slight, non-significant (P≥0.05) upregulation in PIRH2 gene expression was observed in the metformin-treated cells compared to its experimental control. As in the MCF-7 cells, the upregulation in the expression was not dose-dependent.
Like ΔNp73 and PIRH2, MDM2 is a negative regulator of the p53 gene (Fig. 5D). It was observed that the relative expression of the MDM2 gene was 45.6 in the experimental control MCF-7 cells. Treatment with different concentrations of metformin led to downregulation of MDM2 gene expression, however, the dose-dependent downregulation was not much evident. Statistical analysis showed that compared to MCF-7 experimental control, the downregulation in the expression of MDM2 was not significant (P≥0.05) which is indicated by “#” bar in Figure 5D. In the HEK-293 experimental control, the expression of MDM2 was below 0, and metformin treatment slightly and non-significantly (P≥0.05) upregulated the expression of MDM2 in HEK-293 cells. However, the fold change in expression was at the basal level, closer to 1 (Fig. 5D).
DISCUSSION
Metformin is a biguanide used for the treatment of type 2 diabetes mellitus for decades. It is right now the most prescribed anti-hyperglycemic drug used with a good safety profile as it improves glycemic control of the patient without any additional toxicity problem. Epidemiological studies depicted a correlation between T2DM (Type 2 diabetes mellitus) and the incidence of malignancies. Multiple studies linked breast cancer risk with T2DM until, in 2005, the first paper was published that showed reduced breast cancer risk in T2DM patients treated with metformin compared to the patients who were given some other treatment or therapy for diabetes. Since then, research is going on to find the mechanism behind the anti-tumor activity of metformin and its link to reduced risk of breast cancer. Finding the biological mechanisms of the metastatic phenomena is crucial to search for open therapeutic targets for successful interventions (Fares et al., 2020)
Apart from other mechanisms that were proposed to study metformin’s antitumor activity, one important and promising mechanism is its relation to p53. Metformin limits oxidative phosphorylation that results in reduced ATP formation and ultimately AMP level in the cell rises. This AMPK is a key regulator of energy homeostasis of the body, it senses the energy state of the cell and shifts the metabolism of the cell accordingly. That is how metformin carries out its anti-hyperglycemic activity. AMPK phosphorylates the p53 gene as a result p53 attains stabilized structure and becomes activated. P53 being a tumor suppressor gene plays a major role in apoptosis and cellular senescence of the cancer cells. P53 acts in a positive feedback loop with AMPK by inhibiting mTOR to arrest cell progression (Faria et al., 2019).
Until now BrdU assay was carried out to find the anti-proliferative effect of metformin on MCF-7 cells at 10 µM concentration for different time durations 10 h, 24 h, and 72 h and the results showed that the metformin reduced the proliferation of cells in a time-dependent manner (Queiroz et al., 2014).
The effect of metformin on the esophageal squamous carcinoma cell lines was checked to find the anti-tumor role of metformin. It was discovered that metformin could reduce cell proliferation in the case of squamous cancer cells and the rate of inhibition was found to be directly proportional to the concentration of metformin given and time administration. Proving that metformin can be used as a new approach to treat esophageal squamous carcinoma cases (Shao et al., 2019)
Similar anti-proliferative activity of metformin on MCF-7 breast cancer cells was observed in the present study, where a direct dose-dependent reduction in cell viability and proliferation was observed. In the present study, the EC50 of metformin was found to be 22.75µM.
In a study carried out in 2019, cervical cancer cell line HeLa was treated with 60 mM, and 20 mM of metformin, and the expression of p53 and cyclin D1 was measured. The results showed that the dose was sufficient to increase the p53 expression significantly (Yudhani et al., 2019).
Our studies showed an elevated p53 expression when breast cancer cell line MCF-7 were given the treatment of metformin in a range of concentrations (1 µM, 5 µM, 10 µM, 20 µM, 30 µM). It showed that with the increase in the concentration of metformin the expression of p53 was upregulated in the MCF-7 as well as HEK-293 cells. While the upregulation of expression was 1.8, 2.2 and 3.2-fold were observed in 1 µM, 5 µM and 10 µM treated MCF-7 cells, while treatment with 20 µM and 30 µM metformin upregulated the expression to 4.3-fold and 8.4-fold, respectively. A similar trend was observed in the case of HEK-293. Apparently metformin causes decrease in tumor cell proliferation since it phosphorylates and activates p53 which ultimately causes cell cycle arrest and apoptosis in cancer cells.
MDM-2 (murine double minute 2) is a principle negative regulator of the p53 gene and it maintains a low p53 level in the cell by targeting p53 to rapid proteolytic degradation thus inhibiting its transcriptional activity. That’s why MDM-2 is a logical therapeutic target used for cancer treatment and therapies (Noon et al., 2010).
Our findings showed that when given metformin treatment in the concentration of 1 µM, 5 µM, 10 µM, 20 µM and 30 µM to breast cancer cells MCF-7, the rate of expression of MDM-2 was found to be decreasing as the concentration was increased.
Although there is a dose-dependent downregulation, the fold expression values are comparable to each other. In the case of HEK-293, however the fold change in expression was close to 1.
Presently, there is very limited information on the rate of expression of PIRH2 in the metformin-treated breast cancer cell lines. Our findings show that with the increasing metformin concentration the rate of expression of PIRH2 in MCF-7 cell line has downregulated. However, there were slight changes in fold differences that were observed among different concentrations of metformin used. In the case of metformin-treated HEK-293 cells; an opposite trend was observed where a slight, non-significant (P≥0.05) upregulation in PIRH2 gene expression was observed in the metformin-treated cells compared to its experimental control.
As Pirh2 is a negative regulator of p53, downregulation in the expression level of PIRH2 in the case of MCF7-7 after metformin treatment proves that metformin plays a role to inhibit or decrease the expression of cancerous factor Pirh-2 that ultimately enhanced the p53 expression leading to apoptosis and senescence.
ΔNp73 is an oncogene that inhibits p53 activity and sustains cancer cell proliferation by suppressing apoptosis. The expression analysis of ΔNp73 on esophageal squamous carcinoma cells has been done in the past and the results showed a reduced level of ΔNp73 in vivo (Noon et al., 2010).
However, further studies on the p53 pathway, with longer duration of treatment, and more factors involved in the p53 pathway will provide further insights into the mechanism anti-cancer effects of metformin cancer cells.
CONCLUSION
To conclude, by upregulating the expression of the p53 gene and by negatively regulating cancerous interacting partners Mdm-2, Pirh2, and ΔNp73 involved in the p53 pathway, metformin showed anti-cancer properties and it can be used as a target for therapeutic purposes in case of breast cancer. However, further investigation is required to find the exact mode of action of anti-tumor activity of metformin.
Statement of conflict of interest
The authors have declared no conflict of interest.
References
Akram, M., Iqbal, M., Daniyal, M., and Khan, A.U., 2017. Awareness and current knowledge of breast cancer. Biol. Res., 50: 1-23. https://doi.org/10.1186/s40659-017-0140-9
Duan, W., Gao, L., Wu, X., Zhang, Y., Otterson, G.A., and Villalona-Calero, M.A., 2006. Differential response between the p53 ubiquitin protein ligases Pirh2 and MdM2 following DNA damage in human cancer cells. Exp. Cell Res., 312: 3370-3378. https://doi.org/10.1016/j.yexcr.2006.07.005
Fares, J., Fares, M.Y., Khachfe, H.H., Salhab, H.A., and Fares, Y., 2020. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transd. Targ. Ther., 5: 1-17. https://doi.org/10.1038/s41392-020-0134-x
Faria, J., Negalha, G., Azevedo, A., and Martel, F., 2019. Metformin and breast cancer: Molecular targets. J. Mamm. Glan. Biol. Neopl., 24: 111-123. https://doi.org/10.1007/s10911-019-09429-z
García-Cano, J., Sánchez-Tena, S., Sala-Gaston, J., Figueras, A., Viñals, F., Bartrons, R., Ventura, F., and Rosa, J.L., 2020. Regulation of the MDM2-p53 pathway by the ubiquitin ligase HERC2. Mol. Oncol., 14: 69-86. https://doi.org/10.1002/1878-0261.12592
Isleem, R.M., Alzaharna, M.M., and Sharif, F.A., 2020. Synergistic anticancer effect of combining metformin with olive (Olea europaea L.) leaf crude extract on the human breast cancer cell line MCF-7. J. med. Pl., 8: 30-37.
Noon, A.P., Vlatković, N., Polański, R., Maguire, M., Shawki, H., Parsons, K., and Boyd, M.T., 2010. p53 and MDM2 in renal cell carcinoma: Biomarkers for disease progression and future therapeutic targets? Cancer Interdiscipl. Int. J. Am. Cancer Soc., 116: 780-790. https://doi.org/10.1002/cncr.24841
Putri, H.E., Nutho, B., Rungrotmongkol, T., Sritularak, B., Vinayanuwattikun, C., and Chanvorachote, P., 2021. Bibenzyl analogue DS-1 inhibits MDM2-mediated p53 degradation and sensitizes apoptosis in lung cancer cells. Phytomedicine, 85: 153534. https://doi.org/10.1016/j.phymed.2021.153534
Queiroz, E.A., Puukila, S., Eichler, R., Sampaio, S.C., Forsyth, H.L., Lees, S.J., Barbosa, A.M., Dekker, R.F., Fortes, Z.B., and Khaper, N., 2014. Metformin induces apoptosis and cell cycle arrest mediated by oxidative stress, AMPK and FOXO3a in MCF-7 breast cancer cells. PLoS One, 9: e98207. https://doi.org/10.1371/journal.pone.0098207
Ramos, H., Raimundo, L., and Saraiva, L., 2020. p73: From the p53 shadow to a major pharmacological target in anticancer therapy. Pharmacol. Res., 162: 105245. https://doi.org/10.1016/j.phrs.2020.105245
Repetto, G., Del Peso, A., and Zurita, J.L.M., 2008. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Prot., 3: 1125-1131. https://doi.org/10.1038/nprot.2008.75
Shao, K., Wang, L., and Lu, S., 2019. Effect and mechanism of metformin on side population cells of human esophageal squamous cell carcinoma. Cancer Res. Prevent. Treatm., 46: 20-25.
Wang, Z., and Sun, Y., 2010. Targeting p53 for novel anticancer therapy. Transl. Oncol., 3: 1-12. https://doi.org/10.1593/tlo.09250
Yudhani, R.D., Astuti, I., Mustofa, M., Indarto, D., and Muthmainah, M., 2019. Metformin modulates cyclin D1 and P53 expression to inhibit cell proliferation and to induce apoptosis in cervical cancer cell lines. Asian Pac. J. Cancer Prev., 20: 1667. https://www.ncbi.nlm.nih.gov/pubmed/31244286?dopt=Abstract, https://doi.org/10.31557/APJCP.2019.20.6.1667
Zhang, J., Hou, S., You, Z., Li, G., Xu, S., Li, X., Zhang, X., Lei, B., and Pang, D., 2021. Expression and prognostic values of ARID family members in breast cancer. Aging (Albany, NY), 13: 5621. https://doi.org/10.18632/aging.202489
To share on other social networks, click on any share button. What are these?









